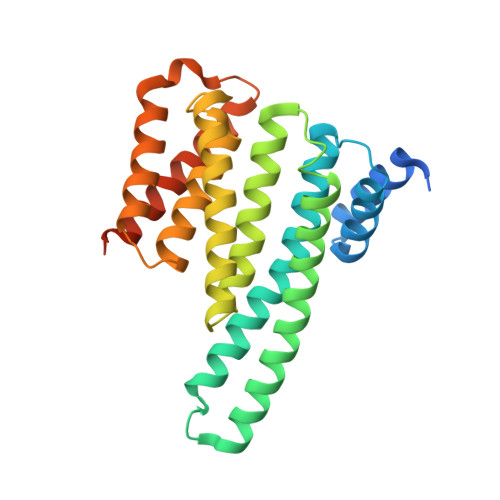
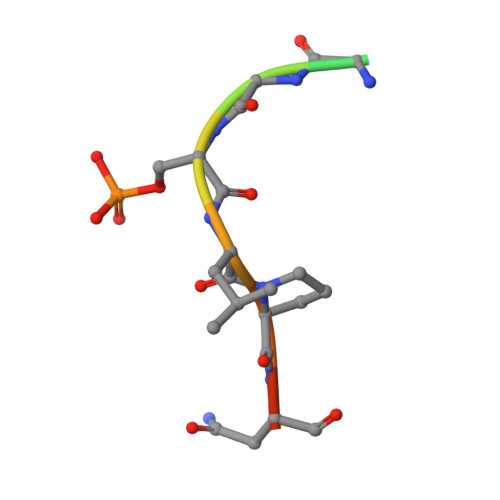
Structural Insights into the Interaction Between CRTCs and 14-3-3.
Chen, H., Zhang, H., Chen, P., Xiang, S.(2021) J Mol Biology 433: 166874-166874
- PubMed: 33556406
- DOI: https://doi.org/10.1016/j.jmb.2021.166874
- Primary Citation of Related Structures:
7D8H, 7D8P, 7D9V - PubMed Abstract:
The CREB-Regulated Transcriptional Coactivators (CRTCs) regulate the transcription of CREB target genes and have important functions in many biological processes. At the basal state, they are phosphorylated at multiple residues, which promotes their association with 14-3-3 that sequesters them in the cytoplasm. Upon dephosphorylation, they translocate into the nuclei and associate with CREB to activate the target gene transcription. Although three conserved serine residues in CRTCs have been implicated in their phosphorylation regulation, whether and how they mediate interactions with 14-3-3 is unclear. Here, we provide direct evidence that these residues and flanking regions interact with 14-3-3 and the structural basis of the interaction. Our study also identified a novel salt bridge in CRTC1 with an important function in binding 14-3-3, expanding the understanding of the interaction between 14-3-3 and its ligands.
- Department of Biochemistry and Molecular Biology, Key Laboratory of Immune Microenvironment and Disease (Ministry of Education), The Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Tianjin Medical University, 300070 Tianjin, PR China.
Organizational Affiliation: