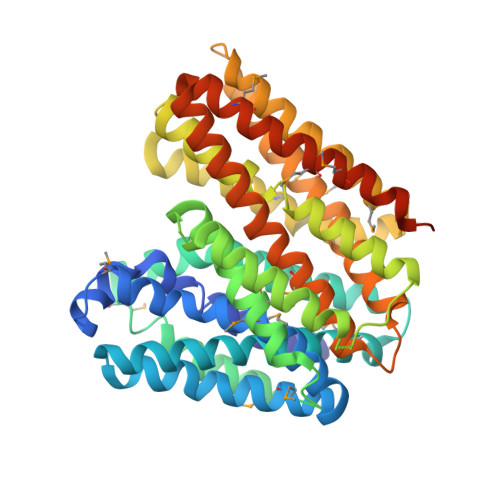
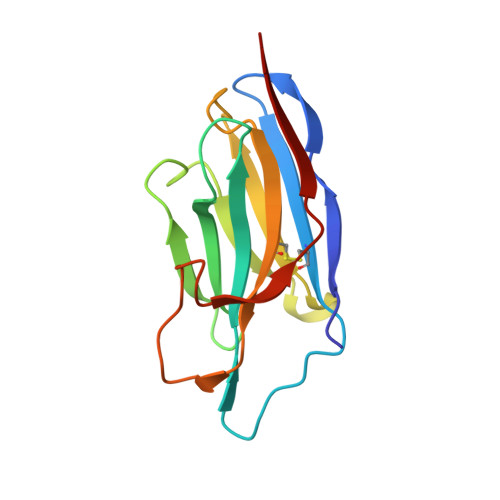
Structural basis of inhibition of a transporter from Staphylococcus aureus, NorC, through a single-domain camelid antibody.
Kumar, S., Athreya, A., Gulati, A., Nair, R.M., Mahendran, I., Ranjan, R., Penmatsa, A.(2021) Commun Biol 4: 836-836
- PubMed: 34226658
- DOI: https://doi.org/10.1038/s42003-021-02357-x
- Primary Citation of Related Structures:
7D5P, 7D5Q - PubMed Abstract:
Transporters play vital roles in acquiring antimicrobial resistance among pathogenic bacteria. In this study, we report the X-ray structure of NorC, a 14-transmembrane major facilitator superfamily member that is implicated in fluoroquinolone resistance in drug-resistant Staphylococcus aureus strains, at a resolution of 3.6 Å. The NorC structure was determined in complex with a single-domain camelid antibody that interacts at the extracellular face of the transporter and stabilizes it in an outward-open conformation. The complementarity determining regions of the antibody enter and block solvent access to the interior of the vestibule, thereby inhibiting alternating-access. NorC specifically interacts with an organic cation, tetraphenylphosphonium, although it does not demonstrate an ability to transport it. The interaction is compromised in the presence of NorC-antibody complex, consequently establishing a strategy to detect and block NorC and related transporters through the use of single-domain camelid antibodies.
- Molecular Biophysics Unit, Indian Institute of Science, Bangalore, India.
Organizational Affiliation: