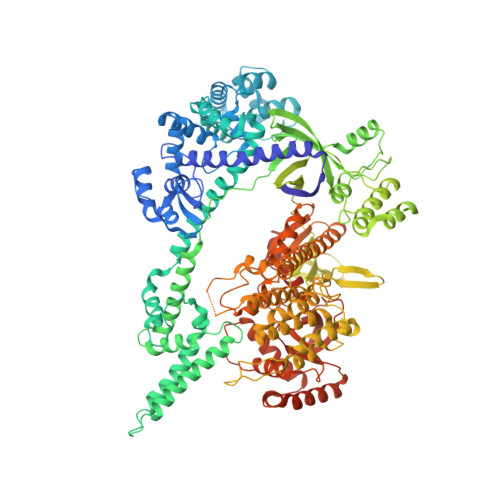
Mechanistic insights into the R-loop formation and cleavage in CRISPR-Cas12i1.
Zhang, B., Luo, D., Li, Y., Perculija, V., Chen, J., Lin, J., Ye, Y., Ouyang, S.(2021) Nat Commun 12: 3476-3476
- PubMed: 34108490
- DOI: https://doi.org/10.1038/s41467-021-23876-5
- Primary Citation of Related Structures:
7D2L, 7D3J, 7D8C, 7EU9 - PubMed Abstract:
Cas12i is a newly identified member of the functionally diverse type V CRISPR-Cas effectors. Although Cas12i has the potential to serve as genome-editing tool, its structural and functional characteristics need to be investigated in more detail before effective application. Here we report the crystal structures of the Cas12i1 R-loop complexes before and after target DNA cleavage to elucidate the mechanisms underlying target DNA duplex unwinding, R-loop formation and cis cleavage. The structure of the R-loop complex after target DNA cleavage also provides information regarding trans cleavage. Besides, we report a crystal structure of the Cas12i1 binary complex interacting with a pseudo target oligonucleotide, which mimics target interrogation. Upon target DNA duplex binding, the Cas12i1 PAM-interacting cleft undergoes a remarkable open-to-closed adjustment. Notably, a zipper motif in the Helical-I domain facilitates unzipping of the target DNA duplex. Formation of the 19-bp crRNA-target DNA strand heteroduplex in the R-loop complexes triggers a conformational rearrangement and unleashes the DNase activity. This study provides valuable insights for developing Cas12i1 into a reliable genome-editing tool.
- The Key Laboratory of Innate Immune Biology of Fujian Province, Provincial University Key Laboratory of Cellular Stress Response and Metabolic Regulation, Biomedical Research Center of South China, Key Laboratory of OptoElectronic Science and Technology for Medicine of Ministry of Education, College of Life Sciences, Fujian Normal University, Fuzhou, China.
Organizational Affiliation: