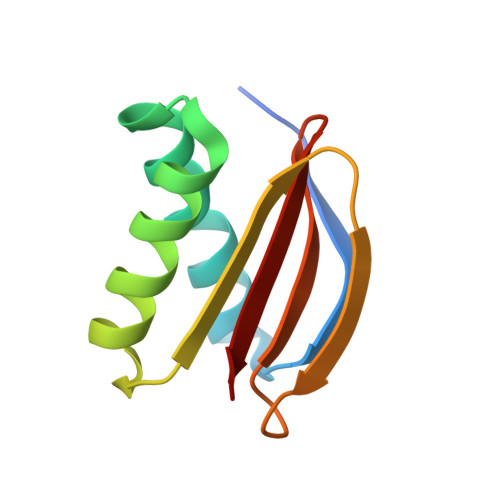
Crystal structure of human LC8 bound to a peptide from Ebola virus VP35.
Lim, D., Shin, H.C., Choi, J.S., Kim, S.J., Ku, B.(2021) J Microbiol 59: 410-416
- PubMed: 33630249
- DOI: https://doi.org/10.1007/s12275-021-0641-7
- Primary Citation of Related Structures:
7D35 - PubMed Abstract:
Zaire ebolavirus, commonly called Ebola virus (EBOV), is an RNA virus that causes severe hemorrhagic fever with high mortality. Viral protein 35 (VP35) is a virulence factor encoded in the EBOV genome. VP35 inhibits host innate immune responses and functions as a critical cofactor for viral RNA replication. EBOV VP35 contains a short conserved motif that interacts with dynein light chain 8 (LC8), which serves as a regulatory hub protein by associating with various LC8-binding proteins. Herein, we present the crystal structure of human LC8 bound to the peptide comprising residues 67-76 of EBOV VP35. Two VP35 peptides were found to interact with homodimeric LC8 by extending the central β-sheets, constituting a 2:2 complex. Structural analysis demonstrated that the intermolecular binding between LC8 and VP35 is mainly sustained by a network of hydrogen bonds and supported by hydrophobic interactions in which Thr73 and Thr75 of VP35 are involved. These findings were verified by binding measurements using isothermal titration calorimetry. Biochemical analyses also verified that residues 67-76 of EBOV VP35 constitute a core region for interaction with LC8. In addition, corresponding motifs from other members of the genus Ebolavirus commonly bound to LC8 but with different binding affinities. Particularly, VP35 peptides originating from pathogenic species interacted with LC8 with higher affinity than those from noninfectious species, suggesting that the binding of VP35 to LC8 is associated with the pathogenicity of the Ebolavirus species.
- Disease Target Structure Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon, 34141, Republic of Korea.
Organizational Affiliation: