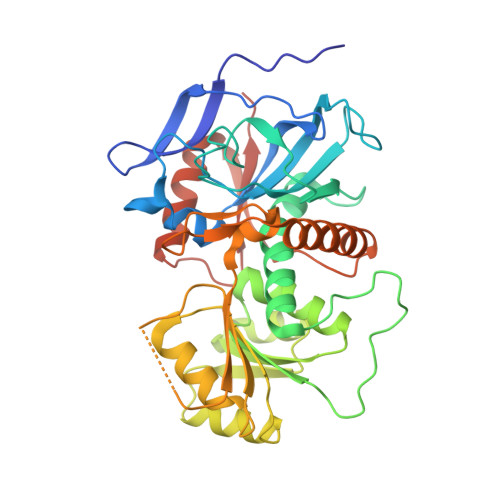
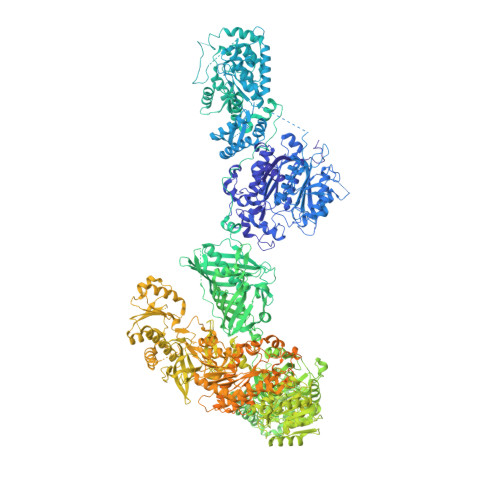
Structural basis for the biosynthesis of lovastatin.
Wang, J., Liang, J., Chen, L., Zhang, W., Kong, L., Peng, C., Su, C., Tang, Y., Deng, Z., Wang, Z.(2021) Nat Commun 12: 867-867
- PubMed: 33558520
- DOI: https://doi.org/10.1038/s41467-021-21174-8
- Primary Citation of Related Structures:
7CPX, 7CPY - PubMed Abstract:
Statins are effective cholesterol-lowering drugs. Lovastatin, one of the precursors of statins, is formed from dihydromonacolin L (DML), which is synthesized by lovastatin nonaketide synthase (LovB), with the assistance of a separate trans-acting enoyl reductase (LovC). A full DML synthesis comprises 8 polyketide synthetic cycles with about 35 steps. The assembling of the LovB-LovC complex, and the structural basis for the iterative and yet permutative functions of the megasynthase have remained a mystery. Here, we present the cryo-EM structures of the LovB-LovC complex at 3.60 Å and the core LovB at 2.91 Å resolution. The domain organization of LovB is an X-shaped face-to-face dimer containing eight connected domains. The binding of LovC laterally to the malonyl-acetyl transferase domain allows the completion of a L-shaped catalytic chamber consisting of six active domains. This architecture and the structural details of the megasynthase provide the basis for the processing of the intermediates by the individual catalytic domains. The detailed architectural model provides structural insights that may enable the re-engineering of the megasynthase for the generation of new statins.
- State Key Laboratory of Microbial Metabolism and School of Life Science & Biotechnology, Shanghai Jiao Tong University, Shanghai, China.
Organizational Affiliation: