Discovery of 6-ethynyl-1H-indole-3-carboxamide Derivatives as Highly Potent and Selective p21-Activated Kinase 4 (PAK4) Inhibitors
Song, P., Zhao, F., Cheng, M.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Serine/threonine-protein kinase PAK 4 | 292 | Homo sapiens | Mutation(s): 0 Gene Names: PAK4, KIAA1142 EC: 2.7.11.1 | 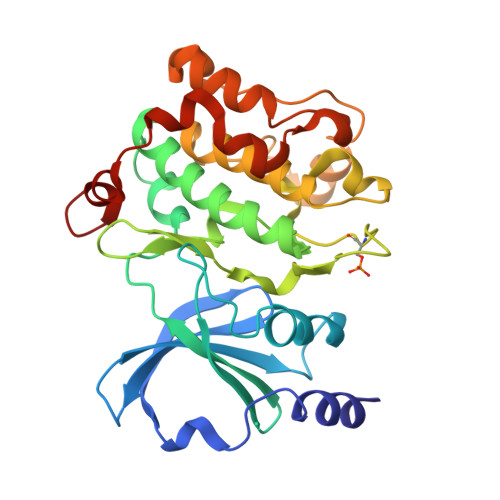 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O96013 (Homo sapiens) Explore O96013 Go to UniProtKB: O96013 | |||||
PHAROS: O96013 GTEx: ENSG00000130669 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O96013 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GC6 (Subject of Investigation/LOI) Query on GC6 | B [auth A] | [(3R)-3-azanylpiperidin-1-yl]-[1-(2-azanylpyrimidin-4-yl)-6-[2-(1-oxidanylcyclohexyl)ethynyl]indol-3-yl]methanone C26 H30 N6 O2 DADSGHOHYQROCR-LJQANCHMSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| SEP Query on SEP | A | L-PEPTIDE LINKING | C3 H8 N O6 P |  | SER |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 64.253 | α = 90 |
| b = 64.253 | β = 90 |
| c = 185.074 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| DENZO | data reduction |
| HKL-2000 | data scaling |
| PDB_EXTRACT | data extraction |
| PHASER | phasing |