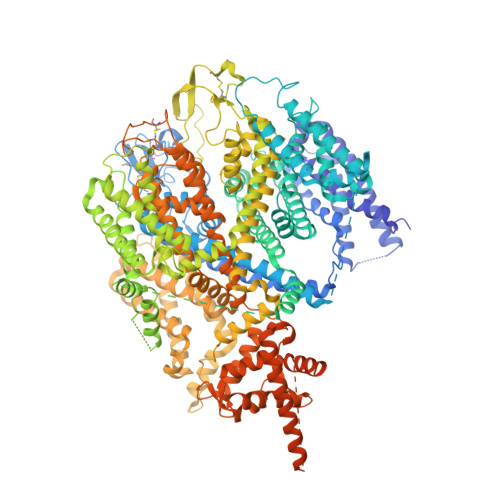
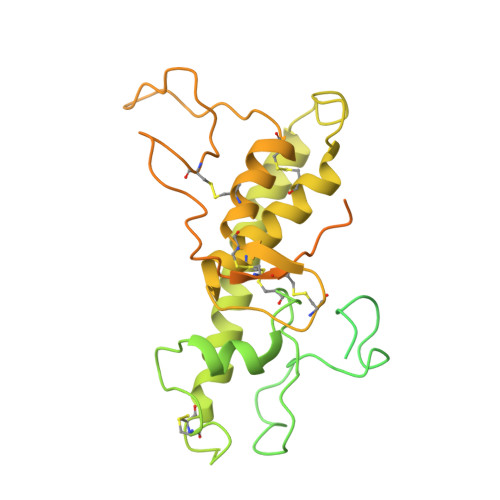
Structure of the human sodium leak channel NALCN in complex with FAM155A.
Xie, J., Ke, M., Xu, L., Lin, S., Huang, J., Zhang, J., Yang, F., Wu, J., Yan, Z.(2020) Nat Commun 11: 5831-5831
- PubMed: 33203861
- DOI: https://doi.org/10.1038/s41467-020-19667-z
- Primary Citation of Related Structures:
7CM3 - PubMed Abstract:
NALCN, a sodium leak channel expressed mainly in the central nervous system, is responsible for the resting Na + permeability that controls neuronal excitability. Dysfunctions of the NALCN channelosome, NALCN with several auxiliary subunits, are associated with a variety of human diseases. Here, we report the cryo-EM structure of human NALCN in complex with FAM155A at an overall resolution of 3.1 angstroms. FAM155A forms extensive interactions with the extracellular loops of NALCN that may help stabilize NALCN in the membrane. A Na + ion-binding site, reminiscent of a Ca 2+ binding site in Ca v channels, is identified in the unique EEKE selectivity filter. Despite its 'leaky' nature, the channel is closed and the intracellular gate is sealed by S6 I , II-III linker and III-IV linker. Our study establishes the molecular basis of Na + permeation and voltage sensitivity, and provides important clues to the mechanistic understanding of NALCN regulation and NALCN channelosome-related diseases.
- Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, 310024, Hangzhou, Zhejiang, China.
Organizational Affiliation: