Crystal structure of the SARS-CoV-2 main protease in complex with Z-VAD(OMe)-FMK
Qiao, J.X., Zeng, R., Wang, Y.F., Li, Y.S., Yao, R., Zhou, Y.L., Chen, P., Lei, J., Yang, S.Y.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 3C-like proteinase | 306 | Severe acute respiratory syndrome coronavirus 2 | Mutation(s): 0 Gene Names: rep, 1a-1b EC: 3.4.22.69 | 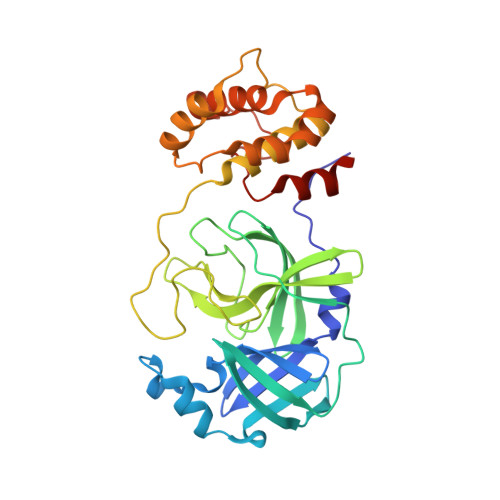 | |
UniProt | |||||
Find proteins for P0DTD1 (Severe acute respiratory syndrome coronavirus 2) Explore P0DTD1 Go to UniProtKB: P0DTD1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0DTD1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Z-VAD(OMe)-FMK | B [auth D] | 5 | synthetic construct | Mutation(s): 0 | 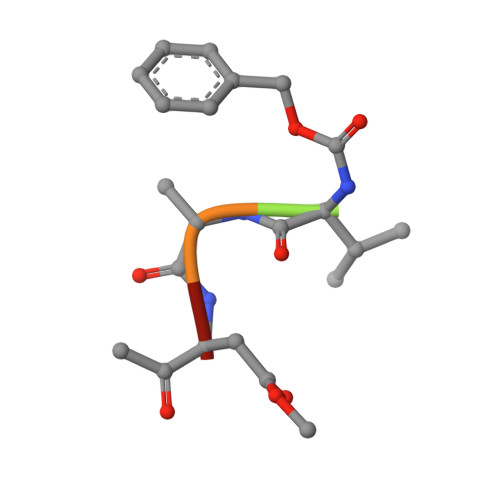 |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CL (Subject of Investigation/LOI) Query on CL | C [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| FL6 Query on FL6 | B [auth D] | L-PEPTIDE LINKING | C5 H9 N O4 |  | ASP |
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| ID | Chains | Name | Type/Class | 2D Diagram | 3D Interactions |
| PRD_002352 Query on PRD_002352 | B [auth D] | Z-VAD(OMe)-FMK | Peptide-like / Inhibitor |  | |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 98.26 | α = 90 |
| b = 82.3 | β = 115.58 |
| c = 51.765 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |
| XDS | data reduction |
| Aimless | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 2020YFS0010/HX2019nCoV039 |