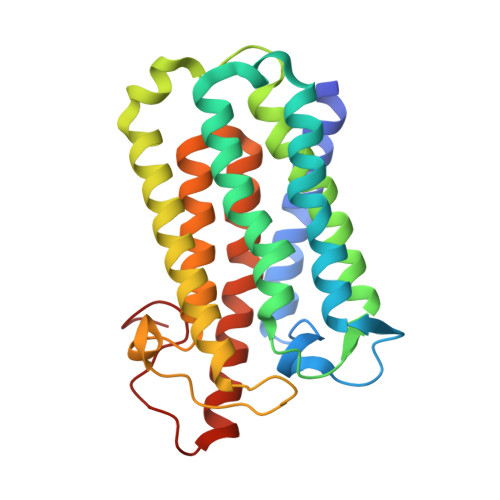
Crystal structure of steroid reductase SRD5A reveals conserved steroid reduction mechanism.
Han, Y., Zhuang, Q., Sun, B., Lv, W., Wang, S., Xiao, Q., Pang, B., Zhou, Y., Wang, F., Chi, P., Wang, Q., Li, Z., Zhu, L., Li, F., Deng, D., Chiang, Y.C., Li, Z., Ren, R.(2021) Nat Commun 12: 449-449
- PubMed: 33469028
- DOI: https://doi.org/10.1038/s41467-020-20675-2
- Primary Citation of Related Structures:
7C83 - PubMed Abstract:
Steroid hormones are essential in stress response, immune system regulation, and reproduction in mammals. Steroids with 3-oxo-Δ 4 structure, such as testosterone or progesterone, are catalyzed by steroid 5α-reductases (SRD5As) to generate their corresponding 3-oxo-5α steroids, which are essential for multiple physiological and pathological processes. SRD5A2 is already a target of clinically relevant drugs. However, the detailed mechanism of SRD5A-mediated reduction remains elusive. Here we report the crystal structure of PbSRD5A from Proteobacteria bacterium, a homolog of both SRD5A1 and SRD5A2, in complex with the cofactor NADPH at 2.0 Å resolution. PbSRD5A exists as a monomer comprised of seven transmembrane segments (TMs). The TM1-4 enclose a hydrophobic substrate binding cavity, whereas TM5-7 coordinate cofactor NADPH through extensive hydrogen bonds network. Homology-based structural models of HsSRD5A1 and -2, together with biochemical characterization, define the substrate binding pocket of SRD5As, explain the properties of disease-related mutants and provide an important framework for further understanding of the mechanism of NADPH mediated steroids 3-oxo-Δ 4 reduction. Based on these analyses, the design of therapeutic molecules targeting SRD5As with improved specificity and therapeutic efficacy would be possible.
- Kobilka Institute of Innovative Drug Discovery, School of Life and Health Sciences, The Chinese University of Hong Kong, Shenzhen, Guangdong, 518172, China.
Organizational Affiliation: