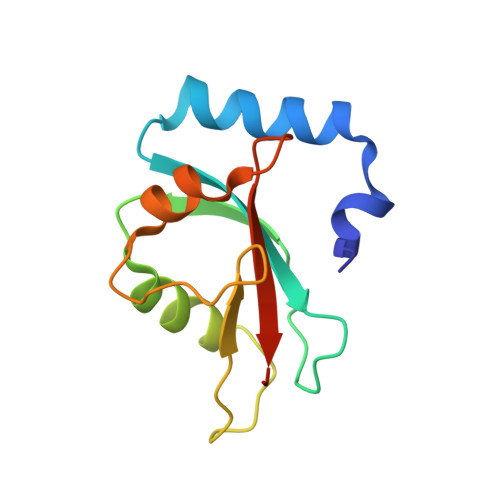
Decoding three distinct states of the Syntaxin17 SNARE motif in mediating autophagosome-lysosome fusion.
Li, Y., Cheng, X., Li, M., Wang, Y., Fu, T., Zhou, Z., Wang, Y., Gong, X., Xu, X., Liu, J., Pan, L.(2020) Proc Natl Acad Sci U S A 117: 21391-21402
- PubMed: 32817423
- DOI: https://doi.org/10.1073/pnas.2006997117
- Primary Citation of Related Structures:
7BV4, 7BV6 - PubMed Abstract:
Syntaxin17, a key autophagosomal N -ethylmaleimide-sensitive factor attachment protein receptor (SNARE) protein, can associate with ATG8 family proteins SNAP29 and VAMP8 to facilitate the membrane fusion process between the double-membraned autophagosome and single-membraned lysosome in mammalian macroautophagy. However, the inherent properties of Syntaxin17 and the mechanistic basis underlying the interactions of Syntaxin17 with its binding proteins remain largely unknown. Here, using biochemical, NMR, and structural approaches, we systemically characterized Syntaxin17 as well as its interactions with ATG8 family proteins, SNAP29 and VAMP8. We discovered that Syntaxin17 alone adopts an autoinhibited conformation mediated by a direct interaction between its Habc domain and the Qa-SNARE motif. In addition, we revealed that the Qa-SNARE region of Syntaxin17 contains one LC3-interacting region (LIR) motif, which preferentially binds to GABARAP subfamily members. Importantly, the GABARAP binding of Syntaxin17 can release its autoinhibited state. The determined crystal structure of the Syntaxin17 LIR-GABARAP complex not only provides mechanistic insights into the interaction between Syntaxin17 and GABARAP but also reveals an unconventional LIR motif with a C-terminally extended 3 10 helix for selectively binding to ATG8 family proteins. Finally, we also elucidated structural arrangements of the autophagic Syntaxin17-SNAP29-VAMP8 SNARE core complex, and uncovered its conserved biochemical and structural characteristics common to all other SNAREs. In all, our findings reveal three distinct states of Syntaxin17, and provide mechanistic insights into the Syntaxin17-mediated autophagosome-lysosome fusion process.
- State Key Laboratory of Bioorganic and Natural Products Chemistry, Center for Excellence in Molecular Synthesis, University of Chinese Academy of Sciences, Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences, 200032 Shanghai, China.
Organizational Affiliation: