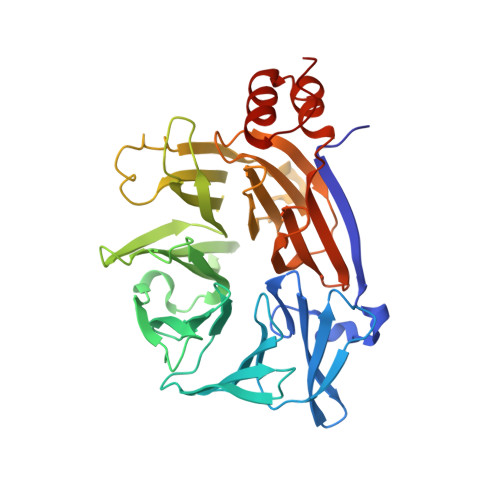
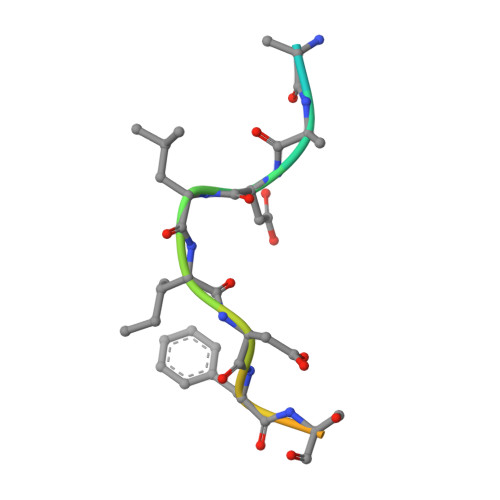
Large-scale phage-based screening reveals extensive pan-viral mimicry of host short linear motifs
Mihalic, F., Simonetti, L., Giudice, G., Sander, M.R., Lindqvist, R., Peters, M.B.A., Benz, C., Kassa, E., Badgujar, D., Inturi, R., Ali, M., Krystkowiak, I., Sayadi, A., Andersson, E., Aronsson, H., Soderberg, O., Dobritzsch, D., Petsalaki, E., Overby, A.K., Jemth, P., Davey, N.E., Ivarsson, Y.(2023) Nat Commun 14