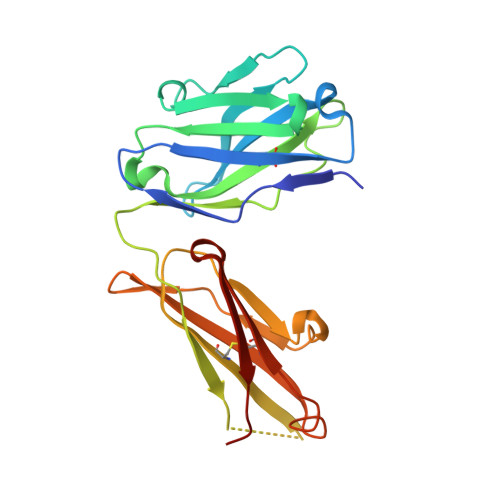
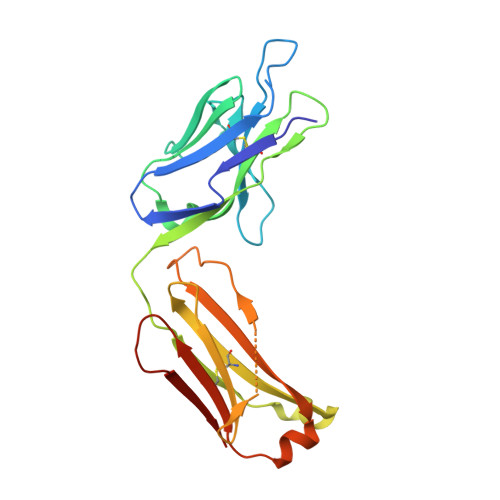
Mass Spectrometry-Based De Novo Sequencing of Monoclonal Antibodies Using Multiple Proteases and a Dual Fragmentation Scheme.
Peng, W., Pronker, M.F., Snijder, J.(2021) J Proteome Res 20: 3559-3566
- PubMed: 34121409
- DOI: https://doi.org/10.1021/acs.jproteome.1c00169
- Primary Citation of Related Structures:
7BG1 - PubMed Abstract:
Antibody sequence information is crucial to understanding the structural basis for antigen binding and enables the use of antibodies as therapeutics and research tools. Here, we demonstrate a method for direct de novo sequencing of monoclonal IgG from the purified antibody products. The method uses a panel of multiple complementary proteases to generate suitable peptides for de novo sequencing by liquid chromatography-tandem mass spectrometry (LC-MS/MS) in a bottom-up fashion. Furthermore, we apply a dual fragmentation scheme, using both stepped high-energy collision dissociation (stepped HCD) and electron-transfer high-energy collision dissociation (EThcD), on all peptide precursors. The method achieves full sequence coverage of the monoclonal antibody herceptin, with an accuracy of 99% in the variable regions. We applied the method to sequence the widely used anti-FLAG-M2 mouse monoclonal antibody, which we successfully validated by remodeling a high-resolution crystal structure of the Fab and demonstrating binding to a FLAG-tagged target protein in Western blot analysis. The method thus offers robust and reliable sequences of monoclonal antibodies.
- Biomolecular Mass Spectrometry and Proteomics, Bijvoet Center for Biomolecular Research and Utrecht Institute of Pharmaceutical Sciences, Utrecht University, Padualaan 8, 3584 CH Utrecht, The Netherlands.
Organizational Affiliation: