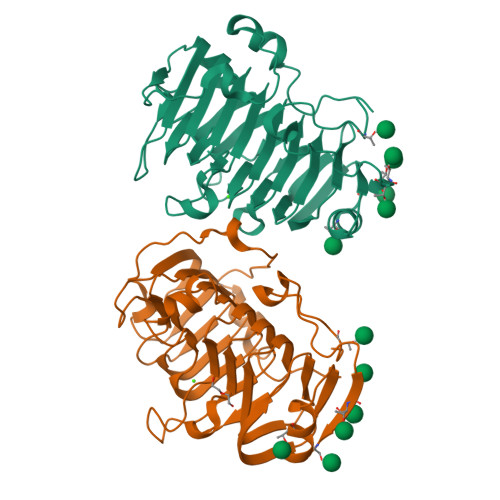
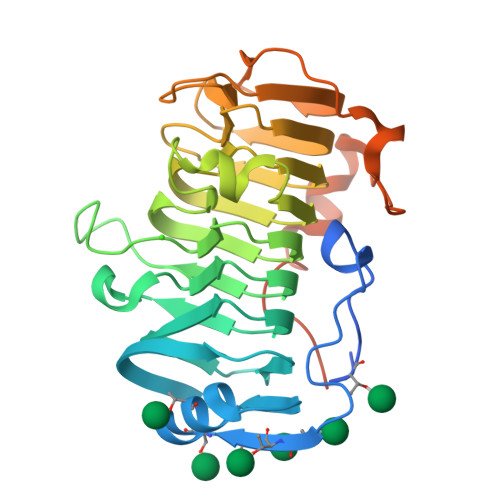
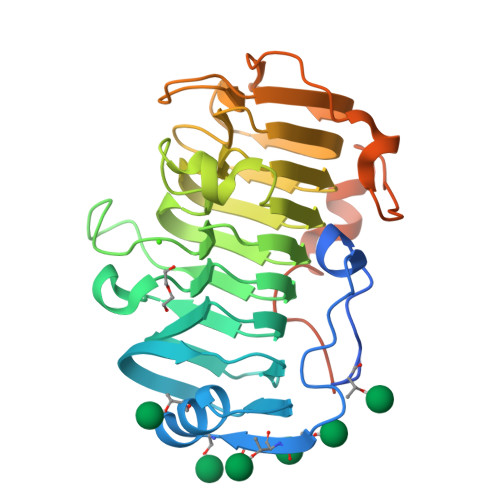
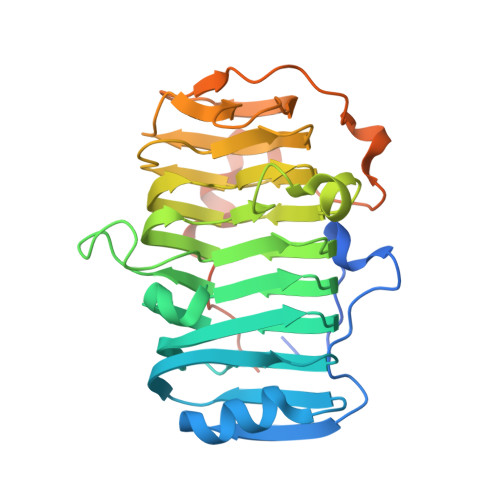
The specificity of pectate lyase VdPelB from Verticilium dahliae is highlighted by structural, dynamical and biochemical characterizations.
Safran, J., Ung, V., Bouckaert, J., Habrylo, O., Molinie, R., Fontaine, J.X., Lemaire, A., Voxeur, A., Pilard, S., Pau-Roblot, C., Mercadante, D., Pelloux, J., Senechal, F.(2023) Int J Biol Macromol 231: 123137-123137
- PubMed: 36639075
- DOI: https://doi.org/10.1016/j.ijbiomac.2023.123137
- Primary Citation of Related Structures:
7BBV - PubMed Abstract:
Pectins, complex polysaccharides and major components of the plant primary cell wall, can be degraded by pectate lyases (PLs). PLs cleave glycosidic bonds of homogalacturonans (HG), the main pectic domain, by β-elimination, releasing unsaturated oligogalacturonides (OGs). To understand the catalytic mechanism and structure/function of these enzymes, we characterized VdPelB from Verticillium dahliae. We first solved the crystal structure of VdPelB at 1.2 Å resolution showing that it is a right-handed parallel β-helix structure. Molecular dynamics (MD) simulations further highlighted the dynamics of the enzyme in complex with substrates that vary in their degree of methylesterification, identifying amino acids involved in substrate binding and cleavage of non-methylesterified pectins. We then biochemically characterized wild type and mutated forms of VdPelB. Pectate lyase VdPelB was most active on non-methylesterified pectins, at pH 8.0 in presence of Ca 2+ ions. The VdPelB-G125R mutant was most active at pH 9.0 and showed higher relative activity compared to native enzyme. The OGs released by VdPelB differed to that of previously characterized PLs, showing its peculiar specificity in relation to its structure. OGs released from Verticillium-partially tolerant and sensitive flax cultivars differed which could facilitate the identification VdPelB-mediated elicitors of defence responses.
Organizational Affiliation:
UMR INRAE 1158 BioEcoAgro - Biologie des Plantes et Innovation, Université de Picardie Jules Verne, UFR des Sciences, 33 Rue St Leu, 80039 Amiens, France.