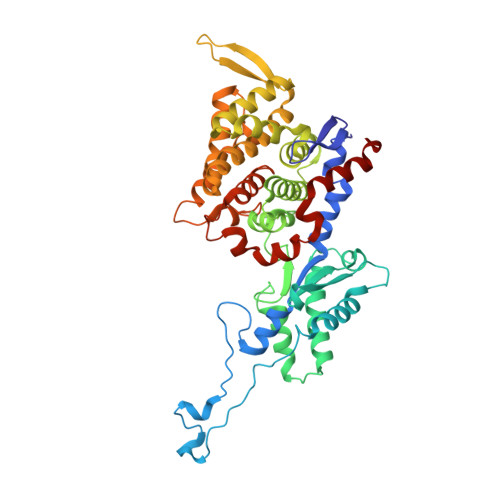
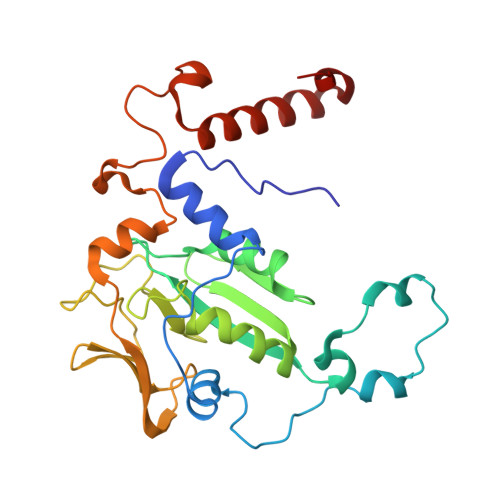
Crystal structure of a key enzyme for anaerobic ethane activation.
Hahn, C.J., Lemaire, O.N., Kahnt, J., Engilberge, S., Wegener, G., Wagner, T.(2021) Science 373: 118-121
- PubMed: 34210888
- DOI: https://doi.org/10.1126/science.abg1765
- Primary Citation of Related Structures:
7B1S, 7B2C, 7B2H - PubMed Abstract:
Ethane, the second most abundant hydrocarbon gas in the seafloor, is efficiently oxidized by anaerobic archaea in syntrophy with sulfate-reducing bacteria. Here, we report the 0.99-angstrom-resolution structure of the proposed ethane-activating enzyme and describe the specific traits that distinguish it from methane-generating and -consuming methyl-coenzyme M reductases. The widened catalytic chamber, harboring a dimethylated nickel-containing F 430 cofactor, would adapt the chemistry of methyl-coenzyme M reductases for a two-carbon substrate. A sulfur from methionine replaces the oxygen from a canonical glutamine as the nickel lower-axial ligand, a feature conserved in thermophilic ethanotrophs. Specific loop extensions, a four-helix bundle dilatation, and posttranslational methylations result in the formation of a 33-angstrom-long hydrophobic tunnel, which guides the ethane to the buried active site as confirmed with xenon pressurization experiments.
- Max Planck Institute for Marine Microbiology, Bremen 28359, Germany.
Organizational Affiliation: