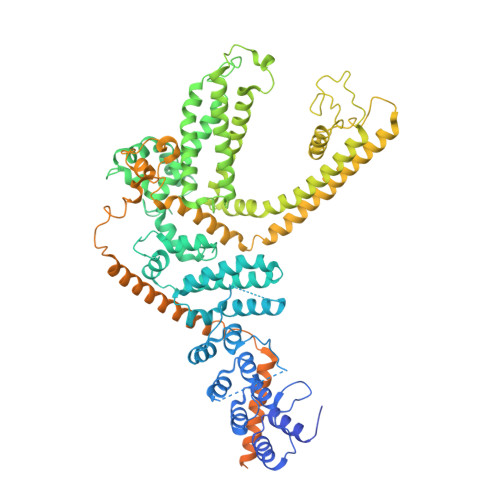
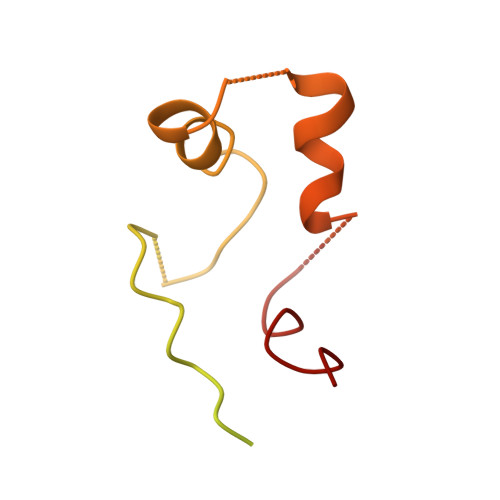
Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Vinayagam, D., Quentin, D., Yu-Strzelczyk, J., Sitsel, O., Merino, F., Stabrin, M., Hofnagel, O., Yu, M., Ledeboer, M.W., Nagel, G., Malojcic, G., Raunser, S.(2020) Elife 9
- PubMed: 33236980
- DOI: https://doi.org/10.7554/eLife.60603
- Primary Citation of Related Structures:
7B05, 7B0J, 7B0S, 7B16, 7B1G - PubMed Abstract:
Canonical transient receptor potential channels (TRPC) are involved in receptor-operated and/or store-operated Ca 2+ signaling. Inhibition of TRPCs by small molecules was shown to be promising in treating renal diseases. In cells, the channels are regulated by calmodulin (CaM). Molecular details of both CaM and drug binding have remained elusive so far. Here, we report structures of TRPC4 in complex with three pyridazinone-based inhibitors and CaM. The structures reveal that all the inhibitors bind to the same cavity of the voltage-sensing-like domain and allow us to describe how structural changes from the ligand-binding site can be transmitted to the central ion-conducting pore of TRPC4. CaM binds to the rib helix of TRPC4, which results in the ordering of a previously disordered region, fixing the channel in its closed conformation. This represents a novel CaM-induced regulatory mechanism of canonical TRP channels.
- Department of Structural Biochemistry, Max Planck Institute of Molecular Physiology, Dortmund, Germany.
Organizational Affiliation: