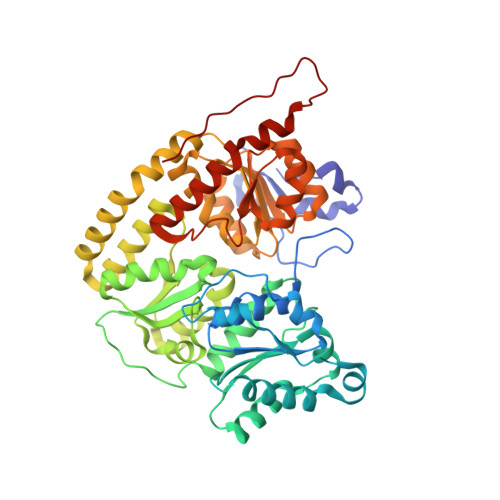
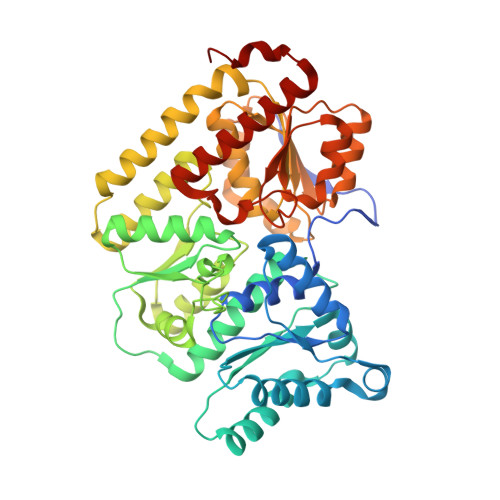
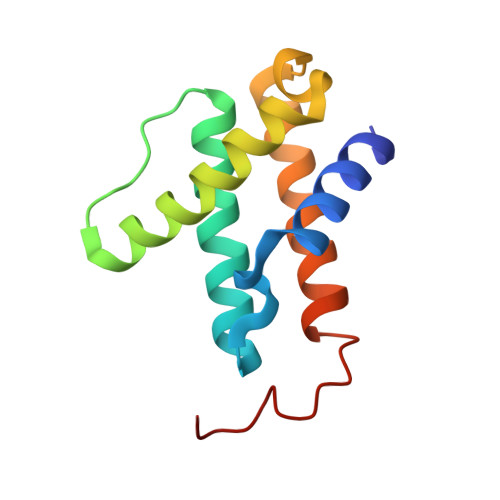
Two ligand-binding sites in CO-reducing V nitrogenase reveal a general mechanistic principle.
Rohde, M., Laun, K., Zebger, I., Stripp, S.T., Einsle, O.(2021) Sci Adv 7
- PubMed: 34049880
- DOI: https://doi.org/10.1126/sciadv.abg4474
- Primary Citation of Related Structures:
7AIZ - PubMed Abstract:
Besides its role in biological nitrogen fixation, vanadium-containing nitrogenase also reduces carbon monoxide (CO) to hydrocarbons, in analogy to the industrial Fischer-Tropsch process. The protein yields 93% of ethylene (C 2 H 4 ), implying a C-C coupling step that mandates the simultaneous binding of two CO at the active site FeV cofactor. Spectroscopic data indicated multiple CO binding events, but structural analyses of Mo and V nitrogenase only confirmed a single site. Here, we report the structure of a two CO-bound state of V nitrogenase at 1.05 Å resolution, with one μ-bridging and one terminal CO molecule. This additional, specific ligand binding site suggests a mechanistic route for CO reduction and hydrocarbon formation, as well as a second access pathway for protons required during the reaction. Moreover, carbonyls are strong-field ligands that are chemically similar to mechanistically relevant hydrides that may be formed and used in a fully analogous fashion.
- Institute for Biochemistry, University of Freiburg, Albertstrasse 21, 79104 Freiburg, Germany.
Organizational Affiliation: