SFX structure of dehaloperoxidase B in the ferric form
Moreno Chicano, T., Carey, L.M., Ebrahim, A., Axford, D.A., Beale, J.H., Sherrell, D.A., Sugimoto, H., Tono, K., Owada, S., Worrall, J.W., Strange, R.W., Owen, R.L., Hough, M.A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Dehaloperoxidase B | 138 | Amphitrite ornata | Mutation(s): 0 | 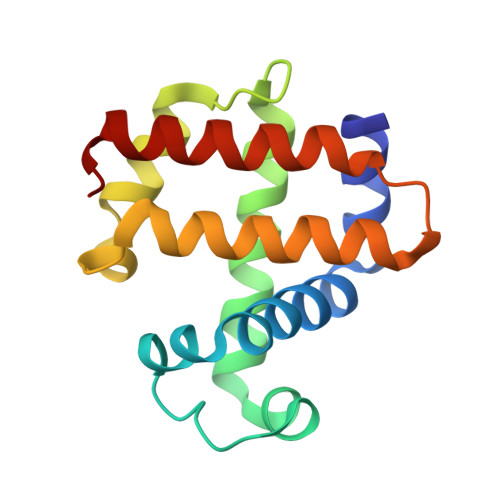 | |
UniProt | |||||
Find proteins for Q9NAV7 (Amphitrite ornata) Explore Q9NAV7 Go to UniProtKB: Q9NAV7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q9NAV7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| HEM Query on HEM | C [auth A], F [auth B] | PROTOPORPHYRIN IX CONTAINING FE C34 H32 Fe N4 O4 KABFMIBPWCXCRK-RGGAHWMASA-L |  | ||
| SO4 Query on SO4 | D [auth A], E [auth A] | SULFATE ION O4 S QAOWNCQODCNURD-UHFFFAOYSA-L |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 61.18 | α = 90 |
| b = 66.995 | β = 90 |
| c = 68.932 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| DIALS | data reduction |
| DIALS | data scaling |
| REFMAC | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Biotechnology and Biological Sciences Research Council (BBSRC) | United Kingdom | BB/R021015/1 |