9.8 MAG. A new host lipid for in meso (lipid cubic phase) crystallization of integral membrane proteins
van Dalsen, L., Smithers, L., Boland, C., Weichert, D., Caffrey, M.(2020) Cryst Growth Des
Experimental Data Snapshot
Starting Model: experimental
View more details
(2020) Cryst Growth Des
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Apolipoprotein N-acyltransferase | 532 | Escherichia coli | Mutation(s): 0 Gene Names: lnt, ACU57_00505, AM464_20560, AUQ13_21565, BMA87_17500, BUE81_17200, BvCms2454_02009, BvCmsHHP001_00880, BvCmsKSNP120_02778, BvCmsKSP076_04015... EC: 2.3.1.269 Membrane Entity: Yes | 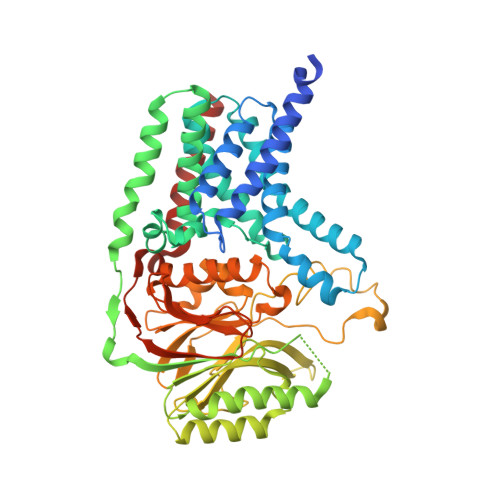 | |
UniProt | |||||
Find proteins for P23930 (Escherichia coli (strain K12)) Explore P23930 Go to UniProtKB: P23930 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P23930 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| LH9 (Subject of Investigation/LOI) Query on LH9 | H [auth A] I [auth A] J [auth A] K [auth A] L [auth A] | [(2~{S})-2,3-bis(oxidanyl)propyl] heptadec-9-enoate C20 H38 O4 NXAQGVNJNZDCNZ-IBGZPJMESA-N |  | ||
| GOL Query on GOL | B [auth A] C [auth A] D [auth A] E [auth A] F [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 48.775 | α = 90 |
| b = 76.173 | β = 90 |
| c = 156.531 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| Coot | model building |
| XDS | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Science Foundation Ireland | Ireland | -- |