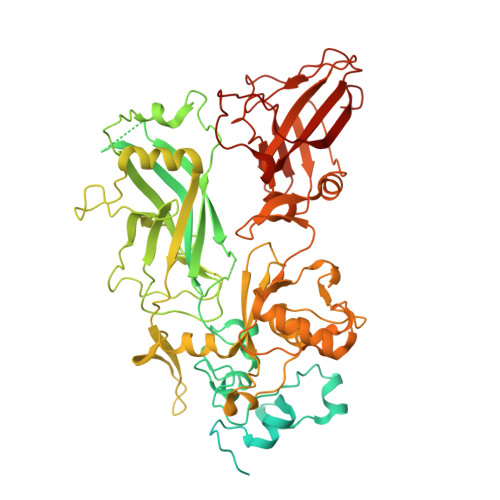
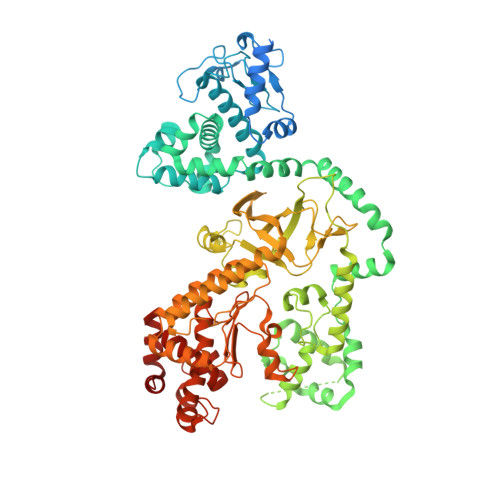
Cryo-EM structure of the fully-loaded asymmetric anthrax lethal toxin in its heptameric pre-pore state.
Antoni, C., Quentin, D., Lang, A.E., Aktories, K., Gatsogiannis, C., Raunser, S.(2020) PLoS Pathog 16: e1008530-e1008530
- PubMed: 32810181
- DOI: https://doi.org/10.1371/journal.ppat.1008530
- Primary Citation of Related Structures:
6ZXJ, 6ZXK, 6ZXL - PubMed Abstract:
Anthrax toxin is the major virulence factor secreted by Bacillus anthracis, causing high mortality in humans and other mammals. It consists of a membrane translocase, known as protective antigen (PA), that catalyzes the unfolding of its cytotoxic substrates lethal factor (LF) and edema factor (EF), followed by translocation into the host cell. Substrate recruitment to the heptameric PA pre-pore and subsequent translocation, however, are not well understood. Here, we report three high-resolution cryo-EM structures of the fully-loaded anthrax lethal toxin in its heptameric pre-pore state, which differ in the position and conformation of LFs. The structures reveal that three LFs interact with the heptameric PA and upon binding change their conformation to form a continuous chain of head-to-tail interactions. As a result of the underlying symmetry mismatch, one LF binding site in PA remains unoccupied. Whereas one LF directly interacts with a part of PA called α-clamp, the others do not interact with this region, indicating an intermediate state between toxin assembly and translocation. Interestingly, the interaction of the N-terminal domain with the α-clamp correlates with a higher flexibility in the C-terminal domain of the protein. Based on our data, we propose a model for toxin assembly, in which the relative position of the N-terminal α-helices in the three LFs determines which factor is translocated first.
- Department of Structural Biochemistry, Max Planck Institute of Molecular Physiology, Dortmund, Germany.
Organizational Affiliation: