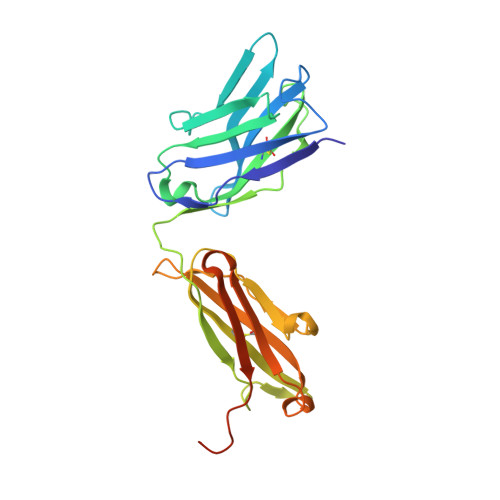
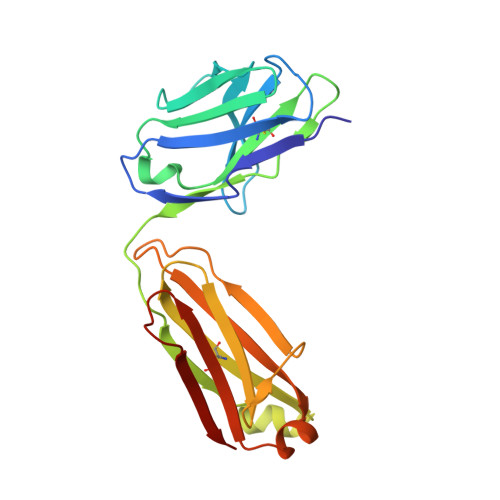
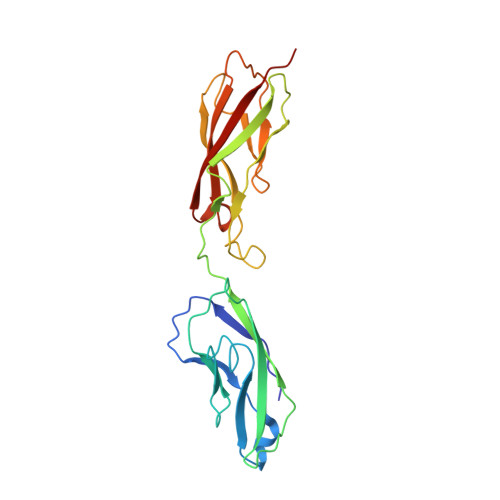
PCA062, a P-cadherin Targeting Antibody-Drug Conjugate, Displays Potent Antitumor Activity Against P-cadherin-expressing Malignancies.
Sheng, Q., D'Alessio, J.A., Menezes, D.L., Karim, C., Tang, Y., Tam, A., Clark, S., Ying, C., Connor, A., Mansfield, K.G., Rondeau, J.M., Ghoddusi, M., Geyer, F.C., Gu, J., McLaughlin, M.E., Newcombe, R., Elliot, G., Tschantz, W.R., Lehmann, S., Fanton, C.P., Miller, K., Huber, T., Rendahl, K.G., Jeffry, U., Pryer, N.K., Lees, E., Kwon, P., Abraham, J.A., Damiano, J.S., Abrams, T.J.(2021) Mol Cancer Ther 20: 1270-1282
- PubMed: 33879555
- DOI: https://doi.org/10.1158/1535-7163.MCT-20-0708
- Primary Citation of Related Structures:
6ZTB, 6ZTF, 6ZTR - PubMed Abstract:
The cell surface glycoprotein P-cadherin is highly expressed in a number of malignancies, including those arising in the epithelium of the bladder, breast, esophagus, lung, and upper aerodigestive system. PCA062 is a P-cadherin specific antibody-drug conjugate that utilizes the clinically validated SMCC-DM1 linker payload to mediate potent cytotoxicity in cell lines expressing high levels of P-cadherin in vitro , while displaying no specific activity in P-cadherin-negative cell lines. High cell surface P-cadherin is necessary, but not sufficient, to mediate PCA062 cytotoxicity. In vivo , PCA062 demonstrated high serum stability and a potent ability to induce mitotic arrest. In addition, PCA062 was efficacious in clinically relevant models of P-cadherin-expressing cancers, including breast, esophageal, and head and neck. Preclinical non-human primate toxicology studies demonstrated a favorable safety profile that supports clinical development. Genome-wide CRISPR screens reveal that expression of the multidrug-resistant gene ABCC1 and the lysosomal transporter SLC46A3 differentially impact tumor cell sensitivity to PCA062. The preclinical data presented here suggest that PCA062 may have clinical value for treating patients with multiple cancer types including basal-like breast cancer.
- Novartis Institutes for Biomedical Research, Cambridge, Massachusetts.
Organizational Affiliation: