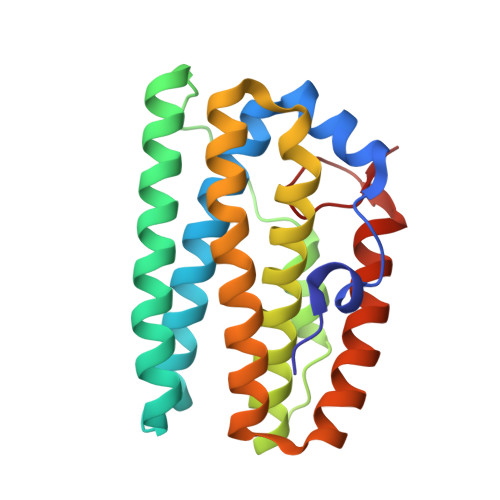
Structural, biochemical and functional analyses of tRNA-monooxygenase enzyme MiaE from Pseudomonas putida provide insights into tRNA/MiaE interaction.
Carpentier, P., Lepretre, C., Basset, C., Douki, T., Torelli, S., Duarte, V., Hamdane, D., Fontecave, M., Atta, M.(2020) Nucleic Acids Res 48: 9918-9930
- PubMed: 32785618
- DOI: https://doi.org/10.1093/nar/gkaa667
- Primary Citation of Related Structures:
6ZMA, 6ZMB, 6ZMC - PubMed Abstract:
MiaE (2-methylthio-N6-isopentenyl-adenosine37-tRNA monooxygenase) is a unique non-heme diiron enzyme that catalyzes the O2-dependent post-transcriptional allylic hydroxylation of a hypermodified nucleotide 2-methylthio-N6-isopentenyl-adenosine (ms2i6A37) at position 37 of selected tRNA molecules to produce 2-methylthio-N6-4-hydroxyisopentenyl-adenosine (ms2io6A37). Here, we report the in vivo activity, biochemical, spectroscopic characterization and X-ray crystal structure of MiaE from Pseudomonas putida. The investigation demonstrates that the putative pp-2188 gene encodes a MiaE enzyme. The structure shows that Pp-MiaE consists of a catalytic diiron(III) domain with a four alpha-helix bundle fold. A docking model of Pp-MiaE in complex with tRNA, combined with site directed mutagenesis and in vivo activity shed light on the importance of an additional linker region for substrate tRNA recognition. Finally, krypton-pressurized Pp-MiaE experiments, revealed the presence of defined O2 site along a conserved hydrophobic tunnel leading to the diiron active center.
- Univ. Grenoble Alpes, CEA, CNRS, CBM-UMR 5249, 17 avenue des martyrs, Grenoble, France.
Organizational Affiliation: