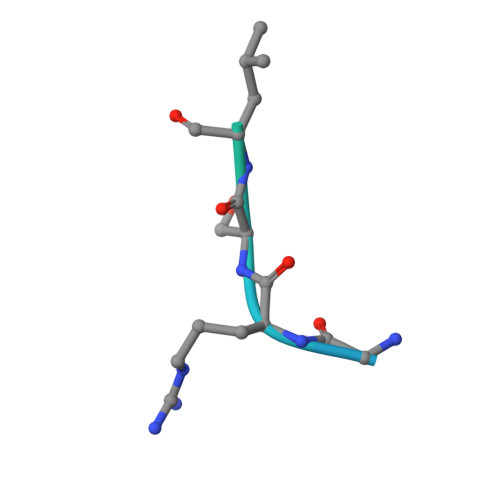
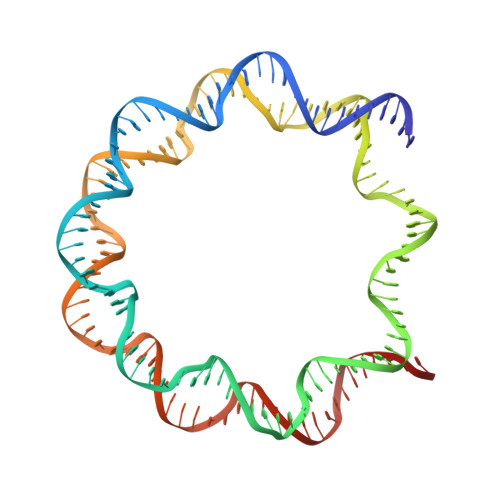
Mechanistic Insights into Regulation of the ALC1 Remodeler by the Nucleosome Acidic Patch.
Lehmann, L.C., Bacic, L., Hewitt, G., Brackmann, K., Sabantsev, A., Gaullier, G., Pytharopoulou, S., Degliesposti, G., Okkenhaug, H., Tan, S., Costa, A., Skehel, J.M., Boulton, S.J., Deindl, S.(2020) Cell Rep 33: 108529-108529
- PubMed: 33357431
- DOI: https://doi.org/10.1016/j.celrep.2020.108529
- Primary Citation of Related Structures:
6ZHX, 6ZHY - PubMed Abstract:
Upon DNA damage, the ALC1/CHD1L nucleosome remodeling enzyme (remodeler) is activated by binding to poly(ADP-ribose). How activated ALC1 recognizes the nucleosome, as well as how this recognition is coupled to remodeling, is unknown. Here, we show that remodeling by ALC1 requires a wild-type acidic patch on the entry side of the nucleosome. The cryo-electron microscopy structure of a nucleosome-ALC1 linker complex reveals a regulatory linker segment that binds to the acidic patch. Mutations within this interface alter the dynamics of ALC1 recruitment to DNA damage and impede the ATPase and remodeling activities of ALC1. Full activation requires acidic patch-linker segment interactions that tether the remodeler to the nucleosome and couple ATP hydrolysis to nucleosome mobilization. Upon DNA damage, such a requirement may be used to modulate ALC1 activity via changes in the nucleosome acidic patches.
- Department of Cell and Molecular Biology, Science for Life Laboratory, Uppsala University, 75124 Uppsala, Sweden.
Organizational Affiliation: