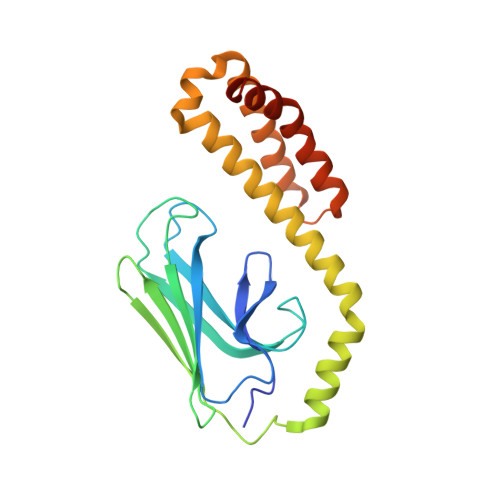
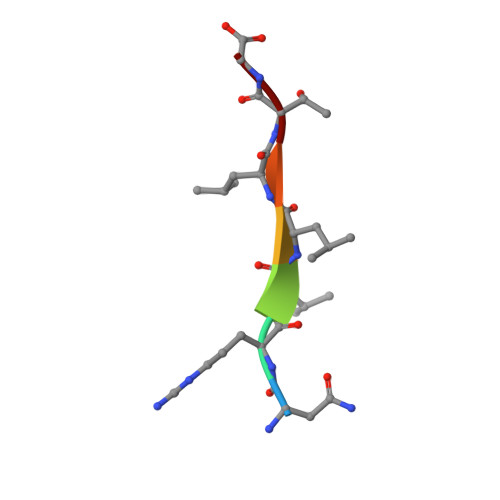
Structure of the substrate-binding domain of Plasmodium falciparum heat-shock protein 70-x.
Schmidt, J., Vakonakis, I.(2020) Acta Crystallogr F Struct Biol Commun 76: 495-500
- PubMed: 33006578
- DOI: https://doi.org/10.1107/S2053230X2001208X
- Primary Citation of Related Structures:
6ZHI - PubMed Abstract:
The malaria parasite Plasmodium falciparum extensively modifies erythrocytes that it invades by exporting a large complement of proteins to the host cell. Among these exported components is a single heat-shock 70 kDa class protein, PfHsp70-x, that supports the virulence and growth rate of the parasite during febrile episodes. The ATP-binding domain of PfHsp70-x has previously been resolved and showed the presence of potentially druggable epitopes that differ from those on human Hsp70 chaperones. Here, the crystallographic structure of the substrate-binding domain (SBD) of PfHsp70-x is presented in complex with a hydrophobic peptide. The PfHsp70-x SBD is shown to be highly similar to the counterpart from a human erythrocytic Hsp70 chaperone. The binding of substrate at the interface between β-sandwich and α-helical subdomains of this chaperone segment is also conserved between the malaria parasite and humans. It is hypothesized that the parasite may partly exploit human chaperones for intra-erythrocytic trafficking and maintenance of its exported proteome.
- Department of Biochemistry, University of Oxford, South Parks Road, Oxford OX1 3QU, United Kingdom.
Organizational Affiliation: