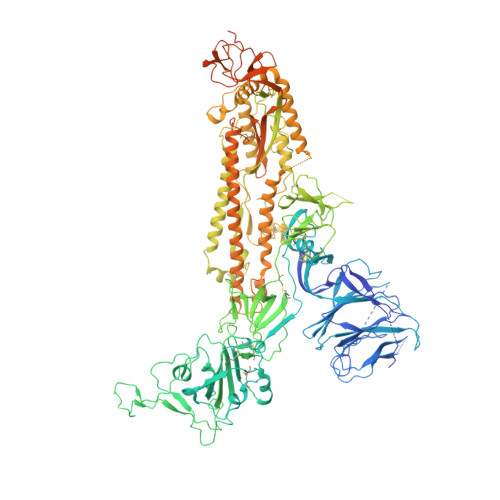
Free fatty acid binding pocket in the locked structure of SARS-CoV-2 spike protein.
Toelzer, C., Gupta, K., Yadav, S.K.N., Borucu, U., Davidson, A.D., Kavanagh Williamson, M., Shoemark, D.K., Garzoni, F., Staufer, O., Milligan, R., Capin, J., Mulholland, A.J., Spatz, J., Fitzgerald, D., Berger, I., Schaffitzel, C.(2020) Science 370: 725-730
- PubMed: 32958580
- DOI: https://doi.org/10.1126/science.abd3255
- Primary Citation of Related Structures:
6ZB4, 6ZB5 - PubMed Abstract:
Coronavirus disease 2019 (COVID-19), caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), represents a global crisis. Key to SARS-CoV-2 therapeutic development is unraveling the mechanisms that drive high infectivity, broad tissue tropism, and severe pathology. Our 2.85-angstrom cryo-electron microscopy structure of SARS-CoV-2 spike (S) glycoprotein reveals that the receptor binding domains tightly bind the essential free fatty acid linoleic acid (LA) in three composite binding pockets. A similar pocket also appears to be present in the highly pathogenic severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV). LA binding stabilizes a locked S conformation, resulting in reduced angiotensin-converting enzyme 2 (ACE2) interaction in vitro . In human cells, LA supplementation synergizes with the COVID-19 drug remdesivir, suppressing SARS-CoV-2 replication. Our structure directly links LA and S, setting the stage for intervention strategies that target LA binding by SARS-CoV-2.
- School of Biochemistry, University of Bristol, 1 Tankard's Close, Bristol BS8 1TD, UK.
Organizational Affiliation: