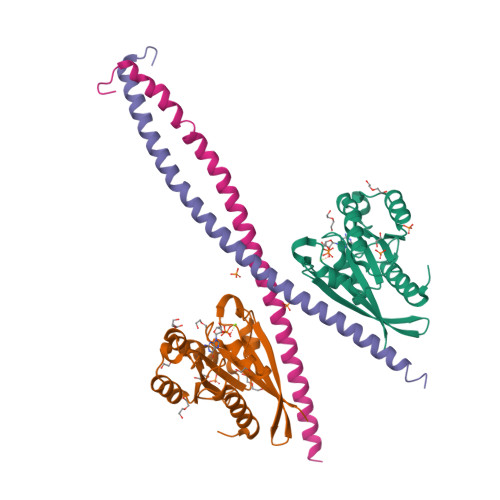
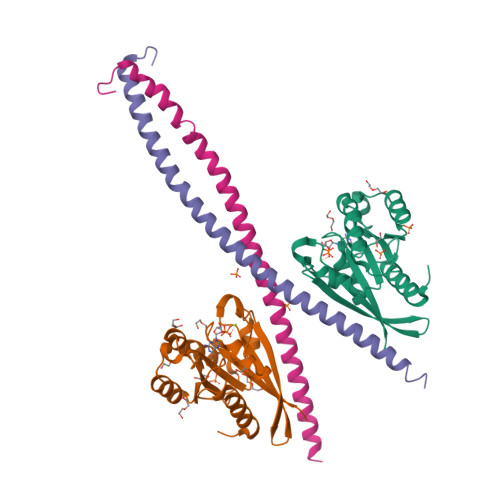
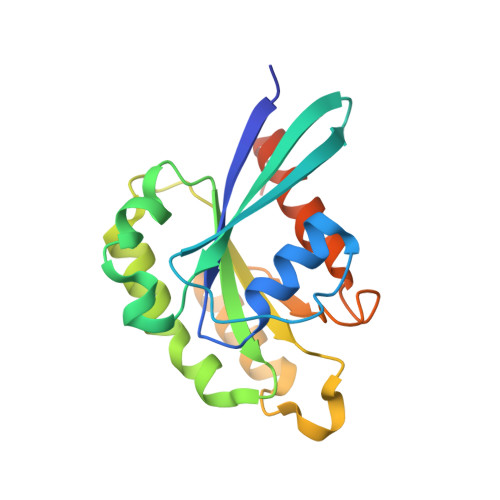
RAB33B recruits the ATG16L1 complex to the phagophore via a noncanonical RAB binding protein.
Pantoom, S., Konstantinidis, G., Voss, S., Han, H., Hofnagel, O., Li, Z., Wu, Y.W.(2021) Autophagy 17: 2290-2304
- PubMed: 32960676
- DOI: https://doi.org/10.1080/15548627.2020.1822629
- Primary Citation of Related Structures:
6Y09, 6ZAY - PubMed Abstract:
Autophagosome formation is a fundamental process in macroautophagy/autophagy, a conserved self-eating mechanism in all eukaryotes, which requires the conjugating ATG (autophagy related) protein complex, ATG12-ATG5-ATG16L1 and lipidated MAP1LC3/LC3 (microtubule associated protein 1 light chain 3). How the ATG12-ATG5-ATG16L1 complex is recruited to membranes is not fully understood. Here, we demonstrated that RAB33B plays a key role in recruiting the ATG16L1 complex to phagophores during starvation-induced autophagy. Crystal structures of RAB33B bound to the coiled-coil domain (CCD) of ATG16L1 revealed the recognition mechanism between RAB33B and ATG16L1. ATG16L1 is a novel RAB-binding protein (RBP) that can induce RAB proteins to adopt active conformation without nucleotide exchange. RAB33B and ATG16L1 mutually determined the localization of each other on phagophores. RAB33B-ATG16L1 interaction was required for LC3 lipidation and autophagosome formation. Upon starvation, a fraction of RAB33B translocated from the Golgi to phagophores and recruited the ATG16L1 complex. In this work, we reported a new mechanism for the recruitment of the ATG12-ATG5-ATG16L1 complex to phagophores by RAB33B, which is required for autophagosome formation. Abbreviations : ATG: autophagy-related; Cα: alpha carbon; CCD: coiled-coil domain; CLEM: correlative light and electron microscopy; DTE: dithioerythritol; EBSS: Earle's balanced salt solution; EDTA: ethylenediaminetetraacetic acid; EGFP: enhanced green fluorescent protein; FBS: fetal bovine serum; FLIM: fluorescence lifetime imaging microscopy; FRET: Förster resonance energy transfer; GDP: guanosine diphosphate; GOLGA2/GM130: golgin A2; GppNHp: guanosine 5'-[β,γ-imido]triphosphate; GST: glutathione S-transferase; GTP: guanosine triphosphate; GTPγS: guanosine 5'-O-[gamma-thio]triphosphate; HA (tag): hemagglutinin (tag); HEK: human embryonic kidney; HeLa: Henrietta Lacks; HEPES: (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid); IgG: immunoglobulin G; K d : dissociation constant; MAP1LC3/LC3: microtubule associated protein 1 light chain 3; MCF7: Michigan cancer foundation-7; MEF: mouse embryonic fibroblast; MEM: minimum essential medium Eagle; MST: microscale thermophoresis; NEAA: non-essential amino acids; PBS: phosphate-buffered saline; PE: phosphatidylethanolamine; PtdIns3P: phosphatidylinositol-3-phosphate; RAB: RAS-associated binding; RB1CC1/FIP200: RB1 inducible coiled-coil protein 1; RBP: RAB-binding protein; SD: standard deviation; SDS: sodium dodecyl sulfate; SQSTM1/p62: sequestosome 1; TBS-T: tris-buffered saline-tween 20; WD (repeat): tryptophan-aspartic acid (repeat); WIPI2B: WD repeat domain phosphoinositide interacting 2B; WT: wild type.
Organizational Affiliation:
Department of Chemistry, Umeå Centre for Microbial Research, Umeå University, Umeå, Sweden.