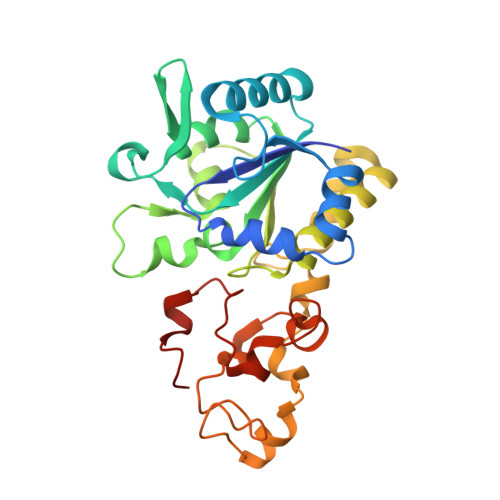
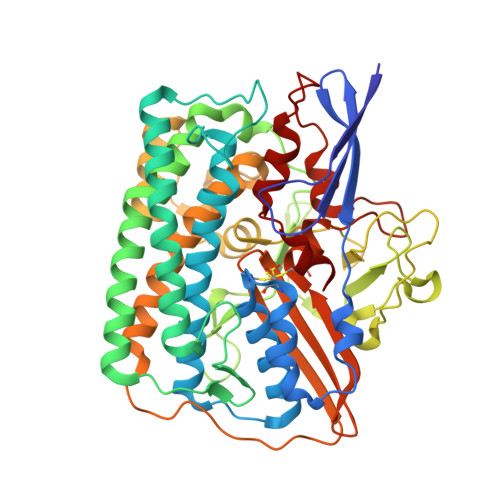
Exploring the gas access routes in a [NiFeSe] hydrogenase using crystals pressurized with krypton and oxygen.
Zacarias, S., Temporao, A., Carpentier, P., van der Linden, P., Pereira, I.A.C., Matias, P.M.(2020) J Biol Inorg Chem 25: 863-874
- PubMed: 32865640
- DOI: https://doi.org/10.1007/s00775-020-01814-y
- Primary Citation of Related Structures:
6Z7R, 6Z8J, 6Z8M, 6Z8O, 6Z9G, 6Z9O, 6ZA1 - PubMed Abstract:
Hydrogenases are metalloenzymes that catalyse both H 2 evolution and uptake. They are gas-processing enzymes with deeply buried active sites, so the gases diffuse through channels that connect the active site to the protein surface. The [NiFeSe] hydrogenases are a special class of hydrogenases containing a selenocysteine as a nickel ligand; they are more catalytically active and less O 2 -sensitive than standard [NiFe] hydrogenases. Characterisation of the channel system of hydrogenases is important to understand how the inhibitor oxygen reaches the active site to cause oxidative damage. To this end, crystals of Desulfovibrio vulgaris Hildenborough [NiFeSe] hydrogenase were pressurized with krypton and oxygen, and a method for tracking labile O 2 molecules was developed, for mapping a hydrophobic channel system similar to that of the [NiFe] enzymes as the major route for gas diffusion.
- Instituto de Tecnologia Química e Biológica António Xavier, Universidade Nova de Lisboa, Av. da República, 2780-157, Oeiras, Portugal.
Organizational Affiliation: