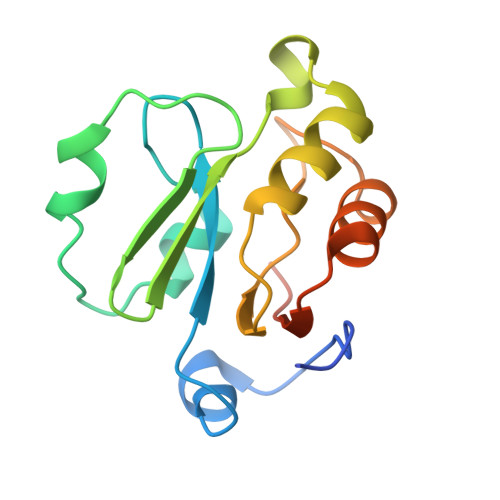
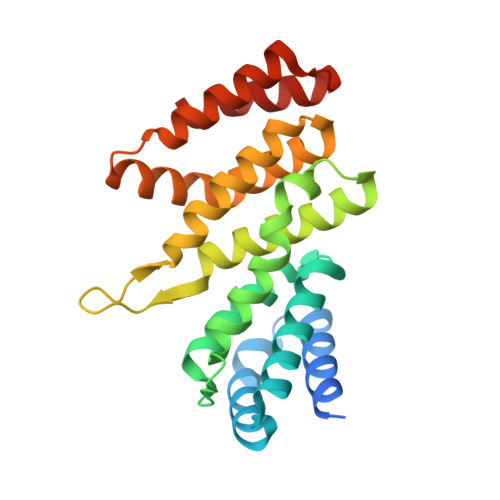
The SARS-unique domain (SUD) of SARS-CoV and SARS-CoV-2 interacts with human Paip1 to enhance viral RNA translation.
Lei, J., Ma-Lauer, Y., Han, Y., Thoms, M., Buschauer, R., Jores, J., Thiel, V., Beckmann, R., Deng, W., Leonhardt, H., Hilgenfeld, R., von Brunn, A.(2021) EMBO J 40: e102277-e102277
- PubMed: 33876849
- DOI: https://doi.org/10.15252/embj.2019102277
- Primary Citation of Related Structures:
6YXJ - PubMed Abstract:
The ongoing outbreak of severe acute respiratory syndrome (SARS) coronavirus 2 (SARS-CoV-2) demonstrates the continuous threat of emerging coronaviruses (CoVs) to public health. SARS-CoV-2 and SARS-CoV share an otherwise non-conserved part of non-structural protein 3 (Nsp3), therefore named as "SARS-unique domain" (SUD). We previously found a yeast-2-hybrid screen interaction of the SARS-CoV SUD with human poly(A)-binding protein (PABP)-interacting protein 1 (Paip1), a stimulator of protein translation. Here, we validate SARS-CoV SUD:Paip1 interaction by size-exclusion chromatography, split-yellow fluorescent protein, and co-immunoprecipitation assays, and confirm such interaction also between the corresponding domain of SARS-CoV-2 and Paip1. The three-dimensional structure of the N-terminal domain of SARS-CoV SUD ("macrodomain II", Mac2) in complex with the middle domain of Paip1, determined by X-ray crystallography and small-angle X-ray scattering, provides insights into the structural determinants of the complex formation. In cellulo, SUD enhances synthesis of viral but not host proteins via binding to Paip1 in pBAC-SARS-CoV replicon-transfected cells. We propose a possible mechanism for stimulation of viral translation by the SUD of SARS-CoV and SARS-CoV-2.
- Institute of Biochemistry, Center for Structural and Cell Biology in Medicine, University of Lübeck, Lübeck, Germany.
Organizational Affiliation: