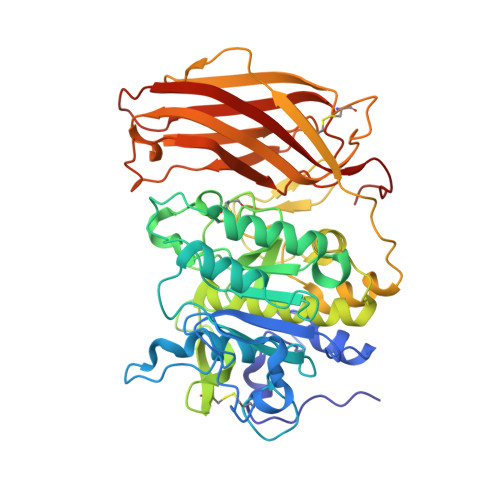
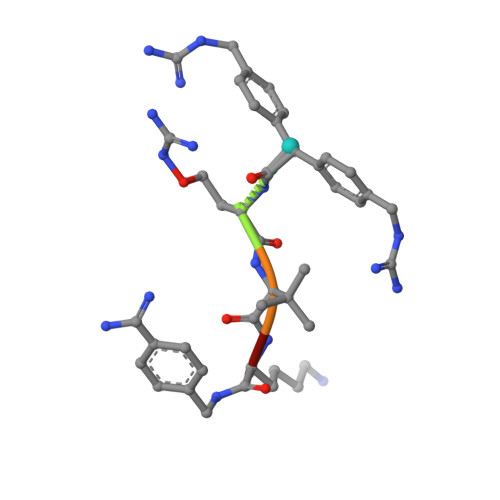
The Basicity Makes the Difference: Improved Canavanine-Derived Inhibitors of the Proprotein Convertase Furin.
Lam van, T.V., Heindl, M.R., Schlutt, C., Bottcher-Friebertshauser, E., Bartenschlager, R., Klebe, G., Brandstetter, H., Dahms, S.O., Steinmetzer, T.(2021) ACS Med Chem Lett 12: 426-432
- PubMed: 33732412
- DOI: https://doi.org/10.1021/acsmedchemlett.0c00651
- Primary Citation of Related Structures:
6YD2, 6YD3, 6YD4, 6YD7 - PubMed Abstract:
Furin activates numerous viral glycoproteins, and its inhibition prevents virus replication and spread. Through the replacement of arginine by the less basic canavanine, new inhibitors targeting furin in the trans-Golgi network were developed. These inhibitors exert potent antiviral activity in cell culture with much lower toxicity than arginine-derived analogues, most likely due to their reduced protonation in the blood circulation. Thus, despite its important physiological functions, furin might be a suitable antiviral drug target.
- Institute of Pharmaceutical Chemistry, Philipps University, Marbacher Weg 6, 35032 Marburg, Germany.
Organizational Affiliation: