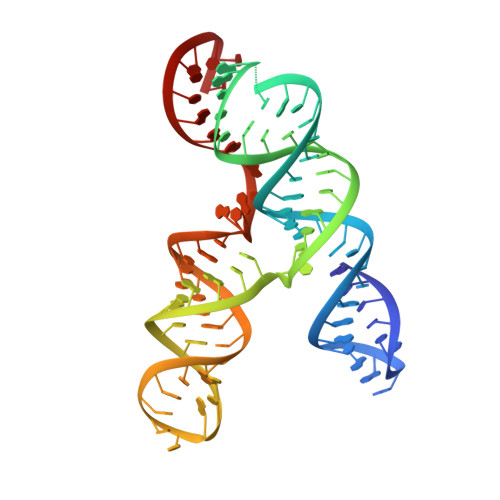
Cryo-electron Microscopy and Exploratory Antisense Targeting of the 28-kDa Frameshift Stimulation Element from the SARS-CoV-2 RNA Genome.
Zhang, K., Zheludev, I.N., Hagey, R.J., Wu, M.T., Haslecker, R., Hou, Y.J., Kretsch, R., Pintilie, G.D., Rangan, R., Kladwang, W., Li, S., Pham, E.A., Bernardin-Souibgui, C., Baric, R.S., Sheahan, T.P., D Souza, V., Glenn, J.S., Chiu, W., Das, R.(2020) bioRxiv
- PubMed: 32743589
- DOI: https://doi.org/10.1101/2020.07.18.209270
- Primary Citation of Related Structures:
6XRZ - PubMed Abstract:
Drug discovery campaigns against Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) are beginning to target the viral RNA genome 1, 2 . The frameshift stimulation element (FSE) of the SARS-CoV-2 genome is required for balanced expression of essential viral proteins and is highly conserved, making it a potential candidate for antiviral targeting by small molecules and oligonucleotides 3-6 . To aid global efforts focusing on SARS-CoV-2 frameshifting, we report exploratory results from frameshifting and cellular replication experiments with locked nucleic acid (LNA) antisense oligonucleotides (ASOs), which support the FSE as a therapeutic target but highlight difficulties in achieving strong inactivation. To understand current limitations, we applied cryogenic electron microscopy (cryo-EM) and the Ribosolve 7 pipeline to determine a three-dimensional structure of the SARS-CoV-2 FSE, validated through an RNA nanostructure tagging method. This is the smallest macromolecule (88 nt; 28 kDa) resolved by single-particle cryo-EM at subnanometer resolution to date. The tertiary structure model, defined to an estimated accuracy of 5.9 Å, presents a topologically complex fold in which the 5' end threads through a ring formed inside a three-stem pseudoknot. Our results suggest an updated model for SARS-CoV-2 frameshifting as well as binding sites that may be targeted by next generation ASOs and small molecules.