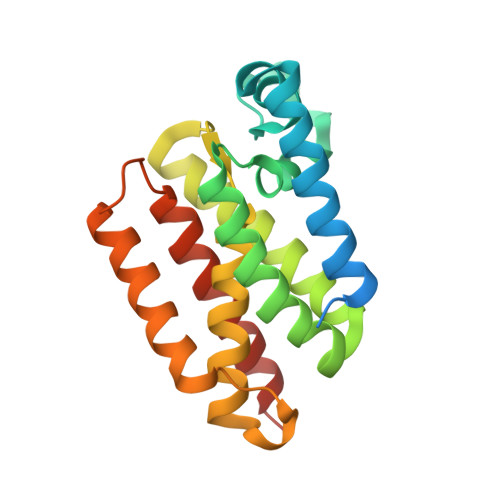
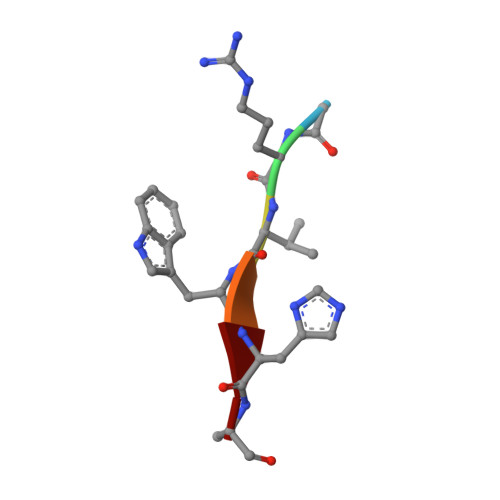
Designed Parasite-Selective Rhomboid Inhibitors Block Invasion and Clear Blood-Stage Malaria.
Gandhi, S., Baker, R.P., Cho, S., Stanchev, S., Strisovsky, K., Urban, S.(2020) Cell Chem Biol 27: 1410-1424.e6
- PubMed: 32888502
- DOI: https://doi.org/10.1016/j.chembiol.2020.08.011
- Primary Citation of Related Structures:
6VJ8, 6VJ9, 6XRO, 6XRP - PubMed Abstract:
Rhomboid intramembrane proteases regulate pathophysiological processes, but their targeting in a disease context has never been achieved. We decoded the atypical substrate specificity of malaria rhomboid PfROM4, but found, unexpectedly, that it results from "steric exclusion": PfROM4 and canonical rhomboid proteases cannot cleave each other's substrates due to reciprocal juxtamembrane steric clashes. Instead, we engineered an optimal sequence that enhanced proteolysis >10-fold, and solved high-resolution structures to discover that boronates enhance inhibition >100-fold. A peptide boronate modeled on our "super-substrate" carrying one "steric-excluding" residue inhibited PfROM4 but not human rhomboid proteolysis. We further screened a library to discover an orthogonal alpha-ketoamide that potently inhibited PfROM4 but not human rhomboid proteolysis. Despite the membrane-immersed target and rapid invasion, ultrastructural analysis revealed that single-dosing blood-stage malaria cultures blocked host-cell invasion and cleared parasitemia. These observations establish a strategy for designing parasite-selective rhomboid inhibitors and expose a druggable dependence on rhomboid proteolysis in non-motile parasites.
- Department of Molecular Biology & Genetics, Johns Hopkins University School of Medicine, Room 507 PCTB, 725 North Wolfe Street, Baltimore, MD 21205, USA.
Organizational Affiliation: