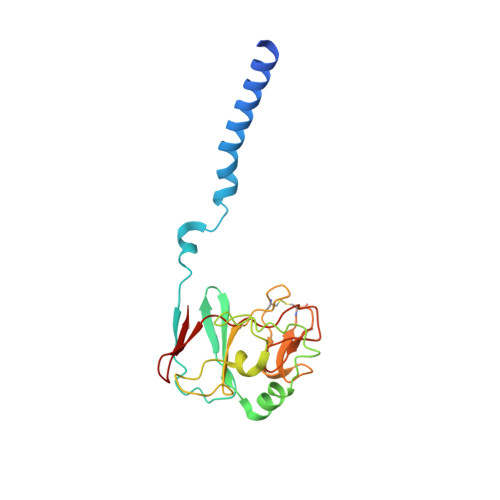
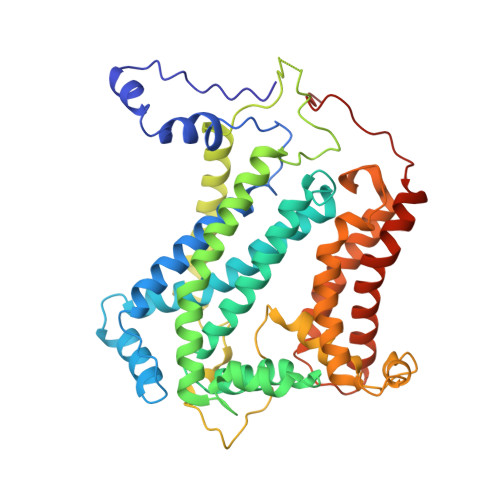
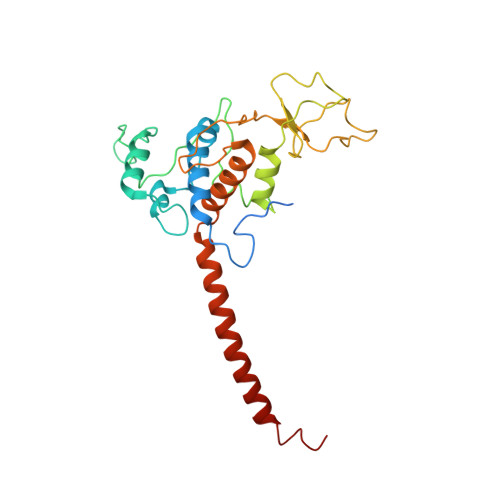
Cryo-EM structures of engineered active bc 1 -cbb 3 type CIII 2 CIV super-complexes and electronic communication between the complexes.
Steimle, S., van Eeuwen, T., Ozturk, Y., Kim, H.J., Braitbard, M., Selamoglu, N., Garcia, B.A., Schneidman-Duhovny, D., Murakami, K., Daldal, F.(2021) Nat Commun 12: 929-929
- PubMed: 33568648
- DOI: https://doi.org/10.1038/s41467-021-21051-4
- Primary Citation of Related Structures:
6XI0, 6XKT, 6XKU, 6XKV, 6XKW, 6XKX, 6XKZ - PubMed Abstract:
Respiratory electron transport complexes are organized as individual entities or combined as large supercomplexes (SC). Gram-negative bacteria deploy a mitochondrial-like cytochrome (cyt) bc 1 (Complex III, CIII 2 ), and may have specific cbb 3 -type cyt c oxidases (Complex IV, CIV) instead of the canonical aa 3 -type CIV. Electron transfer between these complexes is mediated by soluble (c 2 ) and membrane-anchored (c y ) cyts. Here, we report the structure of an engineered bc 1 -cbb 3 type SC (CIII 2 CIV, 5.2 Å resolution) and three conformers of native CIII 2 (3.3 Å resolution). The SC is active in vivo and in vitro, contains all catalytic subunits and cofactors, and two extra transmembrane helices attributed to cyt c y and the assembly factor CcoH. The cyt c y is integral to SC, its cyt domain is mobile and it conveys electrons to CIV differently than cyt c 2 . The successful production of a native-like functional SC and determination of its structure illustrate the characteristics of membrane-confined and membrane-external respiratory electron transport pathways in Gram-negative bacteria.
- Department of Biology, University of Pennsylvania, Philadelphia, PA, USA.
Organizational Affiliation: