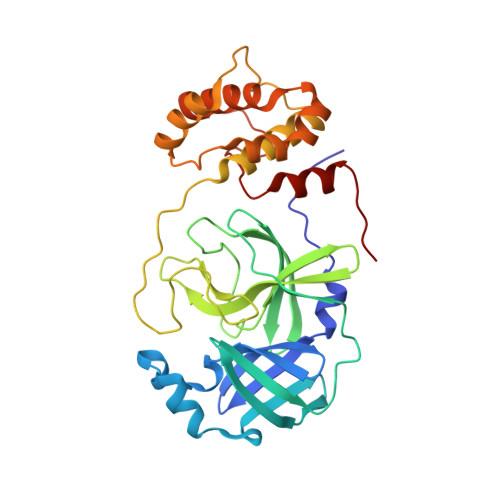
Malleability of the SARS-CoV-2 3CL M pro Active-Site Cavity Facilitates Binding of Clinical Antivirals.
Kneller, D.W., Galanie, S., Phillips, G., O'Neill, H.M., Coates, L., Kovalevsky, A.(2020) Structure 28: 1313
- PubMed: 33152262
- DOI: https://doi.org/10.1016/j.str.2020.10.007
- Primary Citation of Related Structures:
6XCH, 6XQS, 6XQT, 6XQU - PubMed Abstract:
The COVID-19 pandemic caused by SARS-CoV-2 requires rapid development of specific therapeutics and vaccines. The main protease of SARS-CoV-2, 3CL M pro , is an established drug target for the design of inhibitors to stop the virus replication. Repurposing existing clinical drugs can offer a faster route to treatments. Here, we report on the binding mode and inhibition properties of several inhibitors using room temperature X-ray crystallography and in vitro enzyme kinetics. The enzyme active-site cavity reveals a high degree of malleability, allowing aldehyde leupeptin and hepatitis C clinical protease inhibitors (telaprevir, narlaprevir, and boceprevir) to bind and inhibit SARS-CoV-2 3CL M pro . Narlaprevir, boceprevir, and telaprevir are low-micromolar inhibitors, whereas the binding affinity of leupeptin is substantially weaker. Repurposing hepatitis C clinical drugs as COVID-19 treatments may be a useful option to pursue. The observed malleability of the enzyme active-site cavity should be considered for the successful design of specific protease inhibitors.
- Neutron Scattering Division, Oak Ridge National Laboratory, 1 Bethel Valley Road, Oak Ridge, TN 37831, USA; National Virtual Biotechnology Laboratory, US Department of Energy, Washington, DC, USA.
Organizational Affiliation: