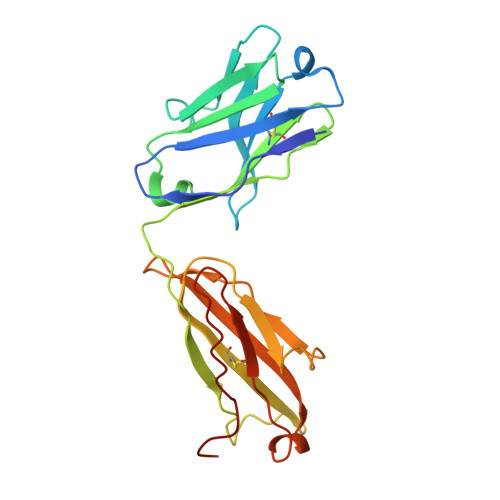
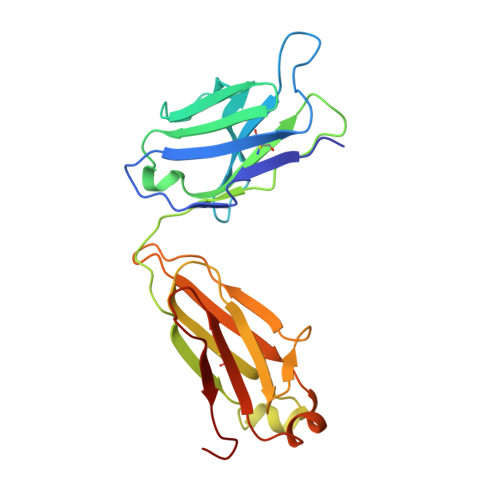
Structural ordering of the Plasmodium berghei circumsporozoite protein repeats by inhibitory antibody 3D11.
Kucharska, I., Thai, E., Srivastava, A., Rubinstein, J.L., Pomes, R., Julien, J.P.(2020) Elife 9
- PubMed: 33253113
- DOI: https://doi.org/10.7554/eLife.59018
- Primary Citation of Related Structures:
6X87, 6X8P, 6X8Q, 6X8S, 6X8U - PubMed Abstract:
Plasmodium sporozoites express circumsporozoite protein (CSP) on their surface, an essential protein that contains central repeating motifs. Antibodies targeting this region can neutralize infection, and the partial efficacy of RTS,S/AS01 - the leading malaria vaccine against P. falciparum (Pf) - has been associated with the humoral response against the repeats. Although structural details of antibody recognition of PfCSP have recently emerged, the molecular basis of antibody-mediated inhibition of other Plasmodium species via CSP binding remains unclear. Here, we analyze the structure and molecular interactions of potent monoclonal antibody (mAb) 3D11 binding to P. berghei CSP (PbCSP) using molecular dynamics simulations, X-ray crystallography, and cryoEM. We reveal that mAb 3D11 can accommodate all subtle variances of the PbCSP repeating motifs, and, upon binding, induces structural ordering of PbCSP through homotypic interactions. Together, our findings uncover common mechanisms of antibody evolution in mammals against the CSP repeats of Plasmodium sporozoites.
- Program in Molecular Medicine, The Hospital for Sick Children Research Institute, Toronto, Canada.
Organizational Affiliation: