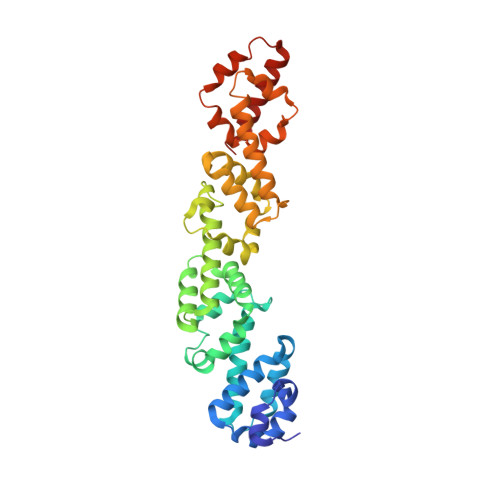Structural and functional study of Legionella pneumophila effector RavA.
Chung, I.Y.W., Li, L., Tyurin, O., Gagarinova, A., Wibawa, R., Li, P., Hartland, E.L., Cygler, M.(2021) Protein Sci 30: 940-955
- PubMed: 33660322
- DOI: https://doi.org/10.1002/pro.4057
- Primary Citation of Related Structures:
6WO6 - PubMed Abstract:
Legionella pneumophila is an intracellular pathogen that causes Legionnaire's disease in humans. This bacterium can be found in freshwater environments as a free-living organism, but it is also an intracellular parasite of protozoa. Human infection occurs when inhaled aerosolized pathogen comes into contact with the alveolar mucosa and replicates in alveolar macrophages. Legionella enters the host cell by phagocytosis and redirects the Legionella-containing phagosomes from the phagocytic maturation pathway. These nascent phagosomes fuse with ER-derived secretory vesicles and membranes forming the Legionella-containing vacuole. Legionella subverts many host cellular processes by secreting over 300 effector proteins into the host cell via the Dot/Icm type IV secretion system. The cellular function for many Dot/Icm effectors is still unknown. Here, we present a structural and functional study of L. pneumophila effector RavA (Lpg0008). Structural analysis revealed that the RavA consists of four ~85 residue long α-helical domains with similar folds, which show only a low level of structural similarity to other protein domains. The ~90 residues long C-terminal segment is predicted to be natively unfolded. We show that during L. pneumophila infection of human cells, RavA localizes to the Golgi apparatus and to the plasma membrane. The same localization is observed when RavA is expressed in human cells. The localization signal resides within the C-terminal sequence C 409 WTSFCGLF 417 . Yeast-two-hybrid screen using RavA as bait identified RAB11A as a potential binding partner. RavA is present in L. pneumophila strains but only distant homologs are found in other Legionella species, where the number of repeats varies.
- Department of Biochemistry, Microbiology and Immunology, University of Saskatchewan, Saskatoon, Saskatchewan, Canada.
Organizational Affiliation: