Caspases from scleractinian coral show unique regulatory features.
Shrestha, S., Tung, J., Grinshpon, R.D., Swartz, P., Hamilton, P.T., Dimos, B., Mydlarz, L., Clark, A.C.(2020) J Biological Chem 295: 14578-14591
- PubMed: 32788218
- DOI: https://doi.org/10.1074/jbc.RA120.014345
- Primary Citation of Related Structures:
6WI4 - PubMed Abstract:
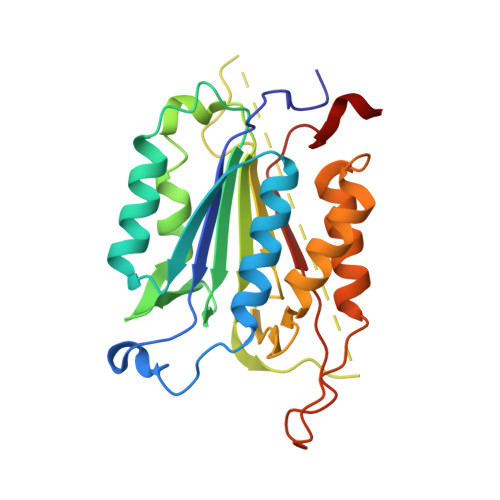
Coral reefs are experiencing precipitous declines around the globe with coral diseases and temperature-induced bleaching being primary drivers of these declines. Regulation of apoptotic cell death is an important component in the coral stress response. Although cnidaria are known to contain complex apoptotic signaling pathways, similar to those in vertebrates, the mechanisms leading to cell death are largely unexplored. We identified and characterized two caspases each from Orbicella faveolata , a disease-sensitive reef-building coral, and Porites astreoides , a disease-resistant reef-building coral. The caspases are predicted homologs of the human executioner caspases-3 and -7, but OfCasp3a ( Orbicella faveolata caspase-3a) and PaCasp7a ( Porites astreoides caspase-7a), which we show to be D XX Dases, contain an N-terminal caspase activation/recruitment domain (CARD) similar to human initiator/inflammatory caspases. OfCasp3b ( Orbicella faveolata caspase-3b) and PaCasp3 ( Porites astreoides caspase-3), which we show to be V XX Dases, have short pro-domains, like human executioner caspases. Our biochemical analyses suggest a mechanism in coral which differs from that of humans, where the CARD-containing D XX Dase is activated on death platforms but the protease does not directly activate the V XX Dase. The first X-ray crystal structure of a coral caspase, of PaCasp7a determined at 1.57 Å resolution, reveals a conserved fold and an N-terminal peptide bound near the active site that may serve as a regulatory exosite. The binding pocket has been observed in initiator caspases of other species. These results suggest mechanisms for the evolution of substrate selection while maintaining common activation mechanisms of CARD-mediated dimerization.
- Department of Biology, University of Texas at Arlington, Arlington, Texas, USA.
Organizational Affiliation: