Structural and biophysical correlation of anti-NANP antibodies with in vivo protection against P. falciparum.
Pholcharee, T., Oyen, D., Flores-Garcia, Y., Gonzalez-Paez, G., Han, Z., Williams, K.L., Volkmuth, W., Emerling, D., Locke, E., Richter King, C., Zavala, F., Wilson, I.A.(2021) Nat Commun 12: 1063-1063
- PubMed: 33594061
- DOI: https://doi.org/10.1038/s41467-021-21221-4
- Primary Citation of Related Structures:
6W00, 6W05, 6WFW, 6WFX, 6WFY, 6WFZ, 6WG0, 6WG1, 6WG2 - PubMed Abstract:
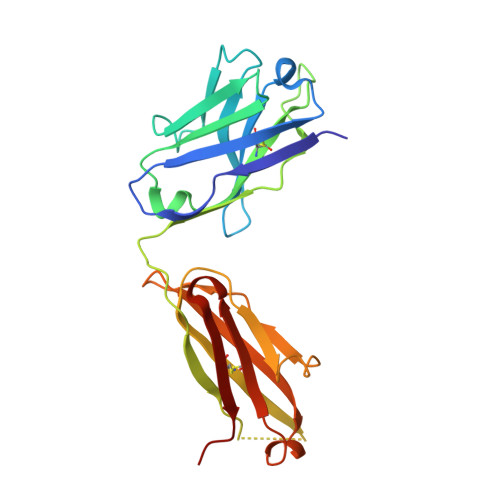
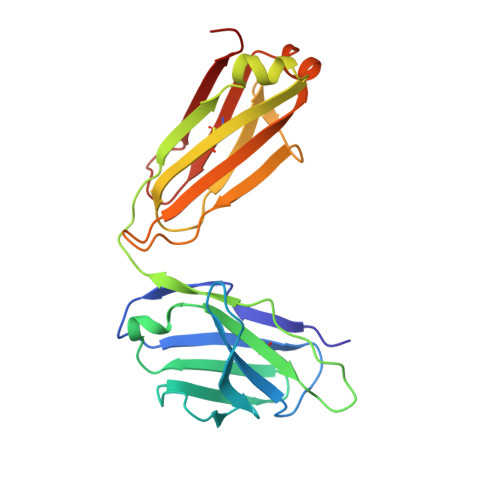
The most advanced P. falciparum circumsporozoite protein-based malaria vaccine, RTS,S/AS01 (RTS,S), confers partial protection but with antibody titers that wane relatively rapidly, highlighting the need to elicit more potent and durable antibody responses. Here, we elucidate crystal structures, binding affinities and kinetics, and in vivo protection of eight anti-NANP antibodies derived from an RTS,S phase 2a trial and encoded by three different heavy-chain germline genes. The structures reinforce the importance of homotypic Fab-Fab interactions in protective antibodies and the overwhelmingly dominant preference for a germline-encoded aromatic residue for recognition of the NANP motif. In this study, antibody apparent affinity correlates best with protection in an in vivo mouse model, with the more potent antibodies also recognizing epitopes with repeating secondary structural motifs of type I β- and Asn pseudo 3 10 turns; such insights can be incorporated into design of more effective immunogens and antibodies for passive immunization.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA, USA.
Organizational Affiliation: