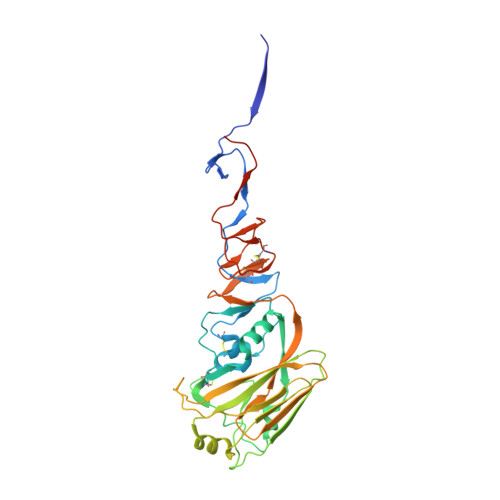
Structure of avian influenza hemagglutinin in complex with a small molecule entry inhibitor.
Antanasijevic, A., Durst, M.A., Cheng, H., Gaisina, I.N., Perez, J.T., Manicassamy, B., Rong, L., Lavie, A., Caffrey, M.(2020) Life Sci Alliance 3
- PubMed: 32611549
- DOI: https://doi.org/10.26508/lsa.202000724
- Primary Citation of Related Structures:
6VMZ - PubMed Abstract:
HA plays a critical role in influenza infection and, thus HA is a potential target for antivirals. Recently, our laboratories have described a novel fusion inhibitor, termed CBS1117, with EC 50 ∼3 μM against group 1 HA. In this work, we characterize the binding properties of CBS1117 to avian H5 HA by x-ray crystallography, NMR, and mutagenesis. The x-ray structure of the complex shows that the compound binds near the HA fusion peptide, a region that plays a critical role in HA-mediated fusion. NMR studies demonstrate binding of CBS1117 to H5 HA in solution and show extensive hydrophobic contacts between the compound and HA surface. Mutagenesis studies further support the location of the compound binding site proximal to the HA fusion peptide and identify additional amino acids that are important to compound binding. Together, this work gives new insights into the CBS1117 mechanism of action and can be exploited to further optimize this compound and better understand the group specific activity of small-molecule inhibitors of HA-mediated entry.
- Department of Biochemistry and Molecular Genetics, University of Illinois at Chicago, Chicago, IL, USA.
Organizational Affiliation: