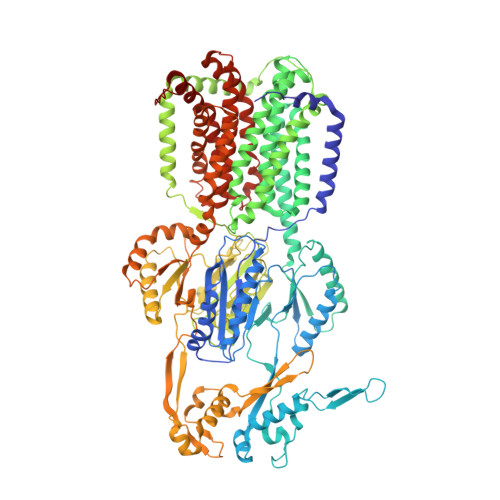
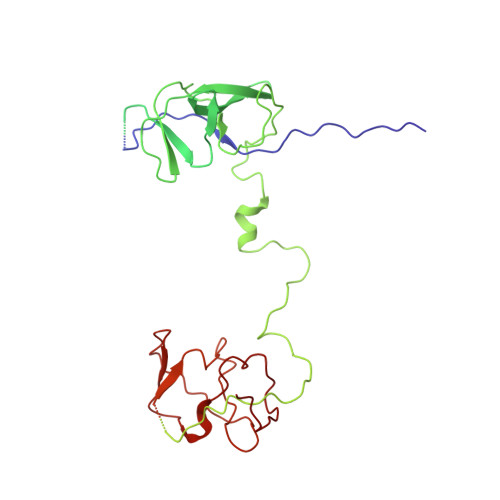
A "Drug Sweeping" State of the TriABC Triclosan Efflux Pump from Pseudomonas aeruginosa.
Fabre, L., Ntreh, A.T., Yazidi, A., Leus, I.V., Weeks, J.W., Bhattacharyya, S., Ruickoldt, J., Rouiller, I., Zgurskaya, H.I., Sygusch, J.(2021) Structure 29: 261
- PubMed: 32966762
- DOI: https://doi.org/10.1016/j.str.2020.09.001
- Primary Citation of Related Structures:
6VEJ - PubMed Abstract:
The structure of the TriABC inner membrane component of the triclosan/SDS-specific efflux pump from Pseudomonas aeruginosa was determined by cryoelectron microscopy to 4.5 Å resolution. The complete structure of the inner membrane transporter TriC of the resistance-nodulation-division (RND) superfamily was solved, including a partial structure of the fused periplasmic membrane fusion subunits, TriA and TriB. The substrate-free conformation of TriABC represents an intermediate step in efflux complex assembly before the engagement of the outer membrane channel. Structural analysis identified a tunnel network whose constriction impedes substrate efflux, indicating inhibition of TriABC in the unengaged state. Blind docking studies revealed binding to TriC at the same loci by substrates and bulkier non-substrates. Together with functional analyses, we propose that selective substrate translocation involves conformational gating at the tunnel narrowing that, together with conformational ordering of TriA and TriB, creates an engaged state capable of mediating substrate efflux.
- McGill University, Department of Anatomy and Cell Biology, Montreal, QC H3A 0G4, Canada.
Organizational Affiliation: