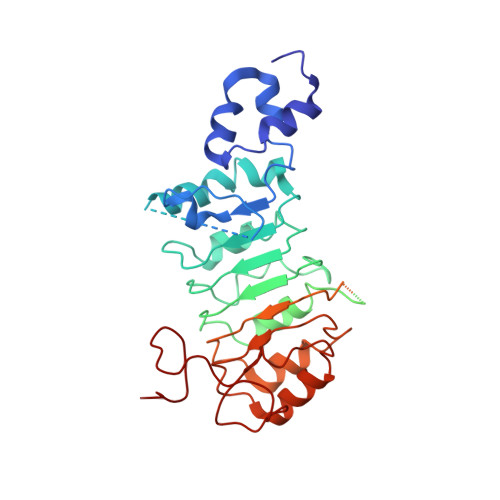
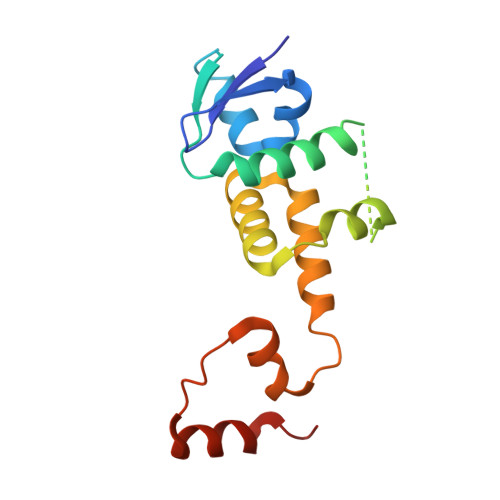
FBXL5 Regulates IRP2 Stability in Iron Homeostasis via an Oxygen-Responsive [2Fe2S] Cluster.
Wang, H., Shi, H., Rajan, M., Canarie, E.R., Hong, S., Simoneschi, D., Pagano, M., Bush, M.F., Stoll, S., Leibold, E.A., Zheng, N.(2020) Mol Cell 78: 31-41.e5
- PubMed: 32126207
- DOI: https://doi.org/10.1016/j.molcel.2020.02.011
- Primary Citation of Related Structures:
6VCD - PubMed Abstract:
Cellular iron homeostasis is dominated by FBXL5-mediated degradation of iron regulatory protein 2 (IRP2), which is dependent on both iron and oxygen. However, how the physical interaction between FBXL5 and IRP2 is regulated remains elusive. Here, we show that the C-terminal substrate-binding domain of FBXL5 harbors a [2Fe2S] cluster in the oxidized state. A cryoelectron microscopy (cryo-EM) structure of the IRP2-FBXL5-SKP1 complex reveals that the cluster organizes the FBXL5 C-terminal loop responsible for recruiting IRP2. Interestingly, IRP2 binding to FBXL5 hinges on the oxidized state of the [2Fe2S] cluster maintained by ambient oxygen, which could explain hypoxia-induced IRP2 stabilization. Steric incompatibility also allows FBXL5 to physically dislodge IRP2 from iron-responsive element RNA to facilitate its turnover. Taken together, our studies have identified an iron-sulfur cluster within FBXL5, which promotes IRP2 polyubiquitination and degradation in response to both iron and oxygen concentrations.
- Department of Pharmacology, University of Washington, Seattle, WA 98195, USA; Howard Hughes Medical Institute, University of Washington, Seattle, WA 98195, USA.
Organizational Affiliation: