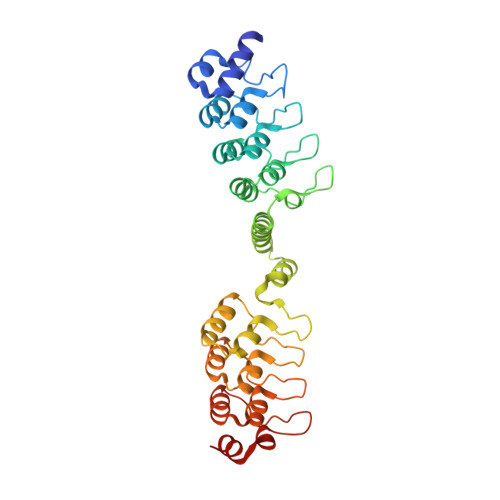
Tissue-Specific Regulation of the Wnt/ beta-Catenin Pathway by PAGE4 Inhibition of Tankyrase.
Koirala, S., Klein, J., Zheng, Y., Glenn, N.O., Eisemann, T., Fon Tacer, K., Miller, D.J., Kulak, O., Lu, M., Finkelstein, D.B., Neale, G., Tillman, H., Vogel, P., Strand, D.W., Lum, L., Brautigam, C.A., Pascal, J.M., Clements, W.K., Potts, P.R.(2020) Cell Rep 32: 107922-107922
- PubMed: 32698014
- DOI: https://doi.org/10.1016/j.celrep.2020.107922
- Primary Citation of Related Structures:
6URQ - PubMed Abstract:
Spatiotemporal control of Wnt/β-catenin signaling is critical for organism development and homeostasis. The poly-(ADP)-ribose polymerase Tankyrase (TNKS1) promotes Wnt/β-catenin signaling through PARylation-mediated degradation of AXIN1, a component of the β-catenin destruction complex. Although Wnt/β-catenin is a niche-restricted signaling program, tissue-specific factors that regulate TNKS1 are not known. Here, we report prostate-associated gene 4 (PAGE4) as a tissue-specific TNKS1 inhibitor that robustly represses canonical Wnt/β-catenin signaling in human cells, zebrafish, and mice. Structural and biochemical studies reveal that PAGE4 acts as an optimal substrate decoy that potently hijacks substrate binding sites on TNKS1 to prevent AXIN1 PARylation and degradation. Consistently, transgenic expression of PAGE4 in mice phenocopies TNKS1 knockout. Physiologically, PAGE4 is selectively expressed in stromal prostate fibroblasts and functions to establish a proper Wnt/β-catenin signaling niche through suppression of autocrine signaling. Our findings reveal a non-canonical mechanism for TNKS1 inhibition that functions to establish tissue-specific control of the Wnt/β-catenin pathway.
- Department of Cell and Molecular Biology, St. Jude Children's Research Hospital, Memphis, TN, USA.
Organizational Affiliation: