Synthetic antibodies against BRIL as universal fiducial marks for single-particle cryoEM structure determination of membrane proteins.
Mukherjee, S., Erramilli, S.K., Ammirati, M., Alvarez, F.J.D., Fennell, K.F., Purdy, M.D., Skrobek, B.M., Radziwon, K., Coukos, J., Kang, Y., Dutka, P., Gao, X., Qiu, X., Yeager, M., Eric Xu, H., Han, S., Kossiakoff, A.A.(2020) Nat Commun 11: 1598-1598
- PubMed: 32221310
- DOI: https://doi.org/10.1038/s41467-020-15363-0
- Primary Citation of Related Structures:
6CBV, 6UR8, 6USF - PubMed Abstract:
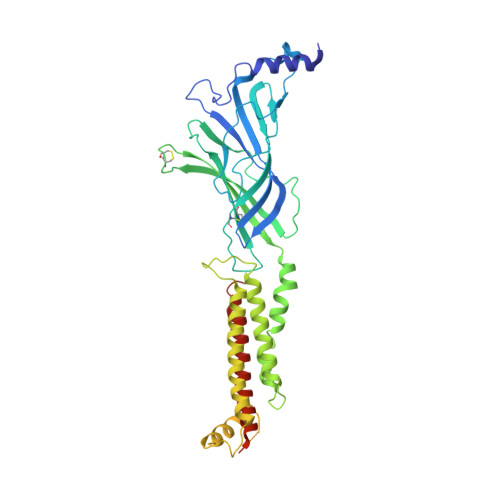
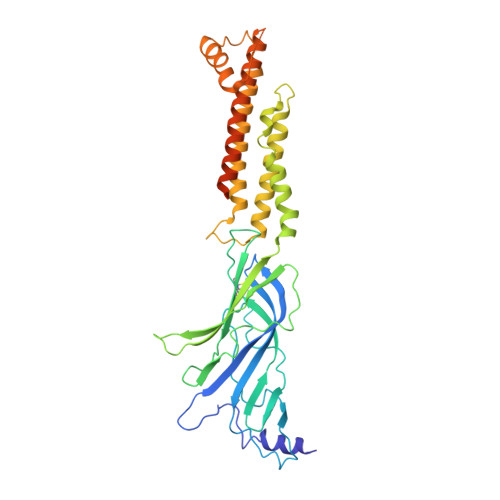
We propose the concept of universal fiducials based on a set of pre-made semi-synthetic antibodies (sABs) generated by customized phage display selections against the fusion protein BRIL, an engineered variant of apocytochrome b562a. These sABs can bind to BRIL fused either into the loops or termini of different GPCRs, ion channels, receptors and transporters without disrupting their structure. A crystal structure of BRIL in complex with an affinity-matured sAB (BAG2) that bound to all systems tested delineates the footprint of interaction. Negative stain and cryoEM data of several examples of BRIL-membrane protein chimera highlight the effectiveness of the sABs as universal fiducial marks. Taken together with a cryoEM structure of sAB bound human nicotinic acetylcholine receptor, this work demonstrates that these anti-BRIL sABs can greatly enhance the particle properties leading to improved cryoEM outcomes, especially for challenging membrane proteins.
- Department of Biochemistry and Molecular Biology, The University of Chicago, Chicago, IL, USA.
Organizational Affiliation: