Structural insights into phosphopantetheinyl hydrolase PptH from Mycobacterium tuberculosis.
Mosior, J., Bourland, R., Soma, S., Nathan, C., Sacchettini, J.(2020) Protein Sci 29: 744-757
- PubMed: 31886928
- DOI: https://doi.org/10.1002/pro.3813
- Primary Citation of Related Structures:
6UNC - PubMed Abstract:
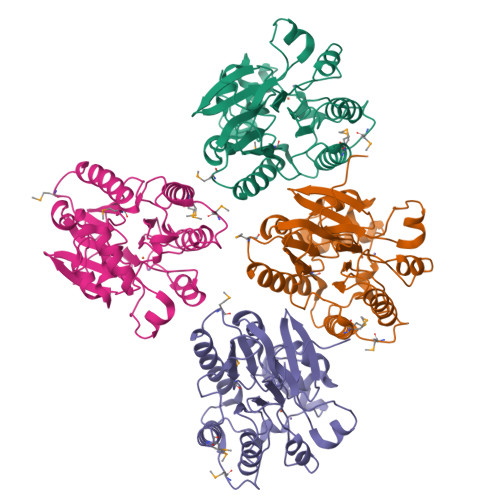
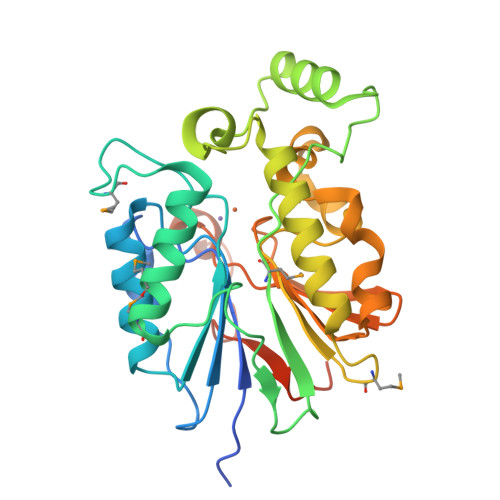
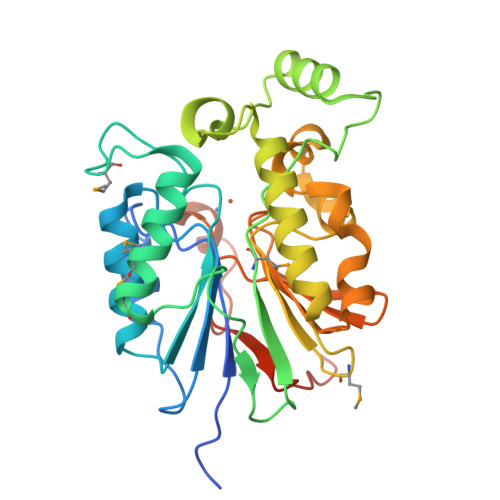
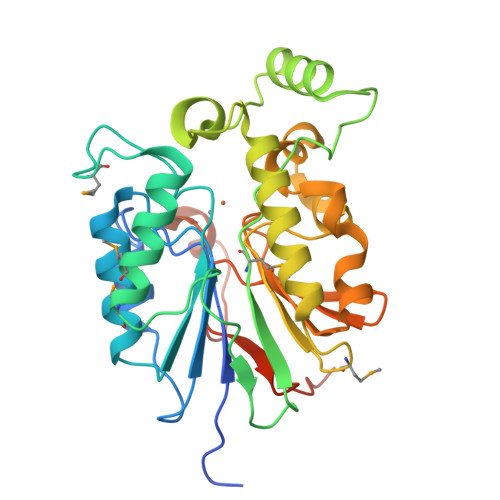
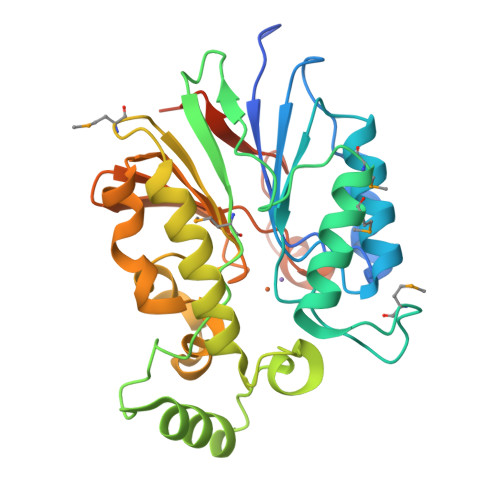
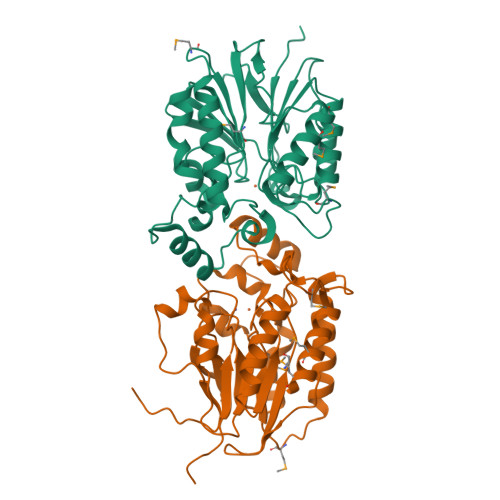
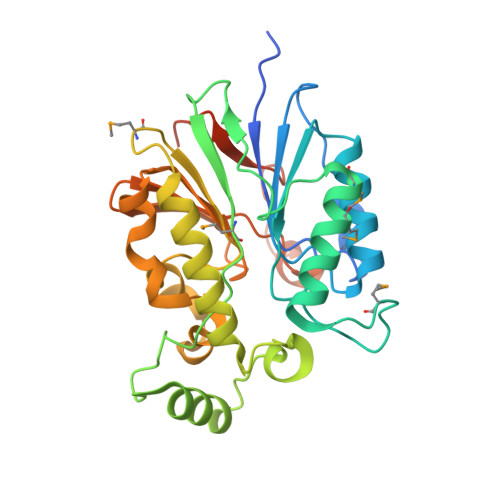
The amidinourea 8918 was recently reported to inhibit the type II phosphopantetheinyl transferase (PPTase) of Mycobacterium tuberculosis (Mtb), PptT, a potential drug-target that activates synthases and synthetases involved in cell wall biosynthesis and secondary metabolism. Surprisingly, high-level resistance to 8918 occurred in Mtb harboring mutations within the gene adjacent to pptT, rv2795c, highlighting the role of the encoded protein as a potentiator of the bactericidal action of the amidinourea. Those studies revealed that Rv2795c (PptH) is a phosphopantetheinyl (PpT) hydrolase, possessing activity antagonistic with respect to PptT. We have solved the crystal structure of Mtb's phosphopantetheinyl hydrolase, making it the first phosphopantetheinyl (carrier protein) hydrolase structurally characterized. The 2.5 Å structure revealed the hydrolases' four-layer (α/β/β/α) sandwich fold featuring a Mn-Fe binuclear center within the active site. A structural similarity search confirmed that PptH most closely resembles previously characterized metallophosphoesterases (MPEs), particularly within the vicinity of the active site, suggesting that it may utilize a similar catalytic mechanism. In addition, analysis of the structure has allowed for the rationalization of the previously reported PptH mutations associated with 8918-resistance. Notably, differences in the sequences and predicted structural characteristics of the PpT hydrolases PptH of Mtb and E. coli's acyl carrier protein hydrolase (AcpH) indicate that the two enzymes evolved convergently and therefore are representative of two distinct PpT hydrolase families.
Organizational Affiliation:
Department of Biochemistry and Biophysics, Texas Agricultural and Mechanical University, College Station, Texas.