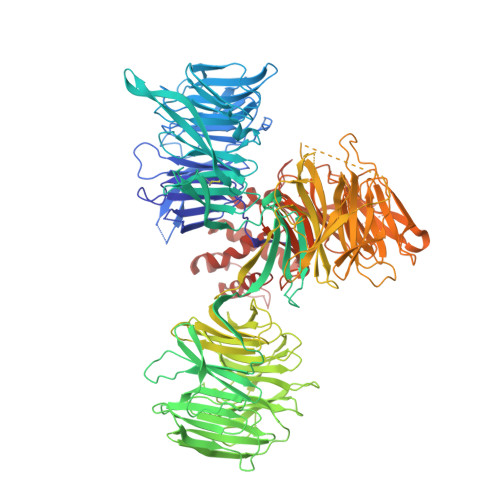
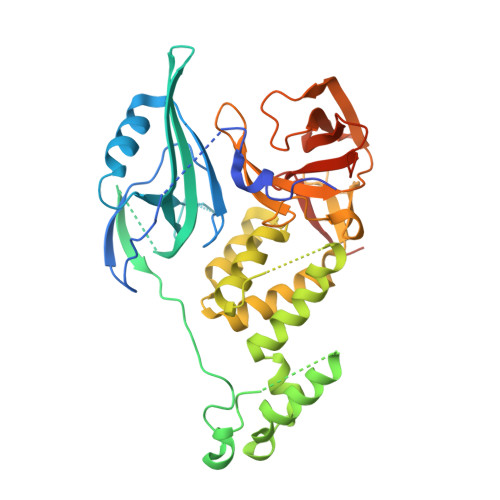
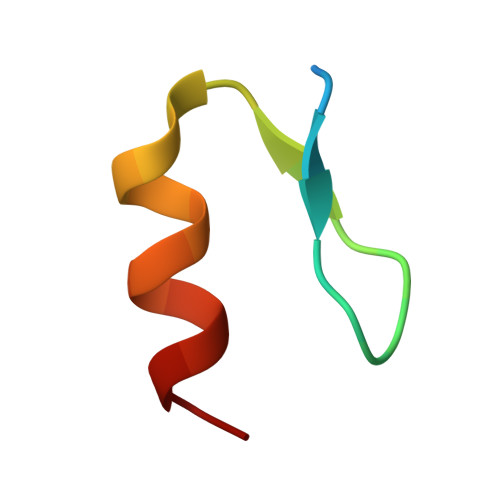
Crystal structure of the SALL4-pomalidomide-cereblon-DDB1 complex.
Matyskiela, M.E., Clayton, T., Zheng, X., Mayne, C., Tran, E., Carpenter, A., Pagarigan, B., McDonald, J., Rolfe, M., Hamann, L.G., Lu, G., Chamberlain, P.P.(2020) Nat Struct Mol Biol 27: 319-322
- PubMed: 32251415
- DOI: https://doi.org/10.1038/s41594-020-0405-9
- Primary Citation of Related Structures:
6UML - PubMed Abstract:
Thalidomide-dependent degradation of the embryonic transcription factor SALL4 by the CRL4 CRBN E3 ubiquitin ligase is a plausible major driver of thalidomide teratogenicity. The structure of the second zinc finger of SALL4 in complex with pomalidomide, cereblon and DDB1 reveals the molecular details of recruitment. Sequence differences and a shifted binding position relative to Ikaros offer a path to the rational design of cereblon-binding drugs with reduced teratogenic risk.
- Celgene Corporation, San Diego, CA, USA. mmatyskiela@celgene.com.
Organizational Affiliation: