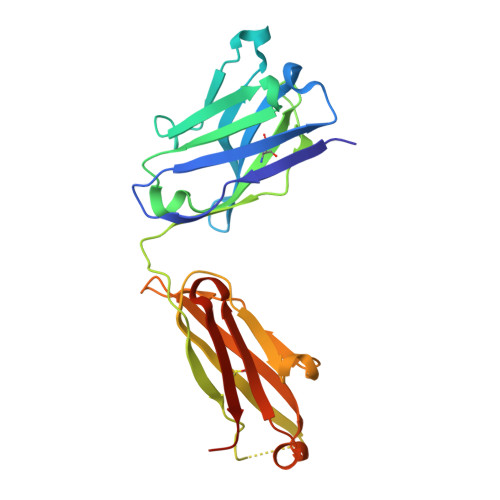
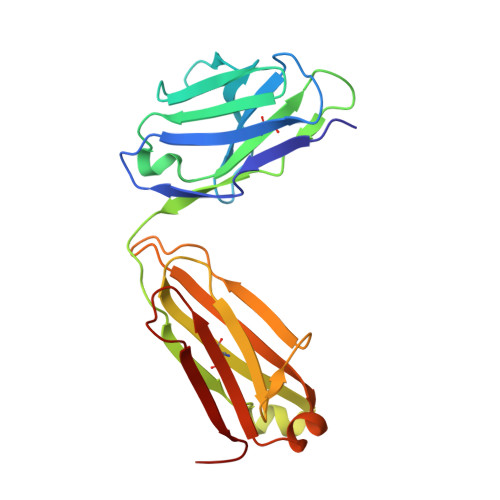
Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
Lerch, T.F., Sharpe, P., Mayclin, S.J., Edwards, T.E., Polleck, S., Rouse, J.C., Zou, Q., Conlon, H.D.(2020) BioDrugs 34: 77-87
- PubMed: 31650490
- DOI: https://doi.org/10.1007/s40259-019-00390-1
- Primary Citation of Related Structures:
6UGS, 6UGT, 6UGU, 6UGV, 6UGW, 6UGX, 6UGY - PubMed Abstract:
Higher-order structure (HOS) assessment is an important component of biosimilarity evaluations. While established spectroscopic methods are routinely used to characterize structure and evaluate similarity, the addition of X-ray crystallographic analysis to these biophysical methods enables orthogonal elucidation of HOS at higher resolution. Crystal structures of the infliximab biosimilar PF-06438179/GP1111 and the reference product Remicade ® , sourced from US and European Union markets, were determined and compared to evaluate HOS similarity. Analytical ultracentrifugation studies were conducted to understand reversible self-association. In contrast to more routine spectroscopic methods, the crystal structures enable three-dimensional assessment of complementarity-determining regions and other local regions at near-atomic resolution. The biosimilar structures are highly similar to those of the reference product, as demonstrated visually and though all-atom root-mean-squared deviation measurements. The structures provide new insights into the physicochemical properties of the proposed biosimilar and the reference product, further strengthening the 'totality of evidence' in the evaluation of similarity.
- Analytical Research and Development, Biotherapeutics Pharmaceutical Sciences, Pfizer Inc., Chesterfield, MO, 63017, USA. thomas.lerch@pfizer.com.
Organizational Affiliation: