Spatially remote motifs cooperatively affect substrate preference of a ruminal GH26-type endo-beta-1,4-mannanase.
Mandelli, F., de Morais, M.A.B., de Lima, E.A., Oliveira, L., Persinoti, G.F., Murakami, M.T.(2020) J Biological Chem 295: 5012-5021
- PubMed: 32139511
- DOI: https://doi.org/10.1074/jbc.RA120.012583
- Primary Citation of Related Structures:
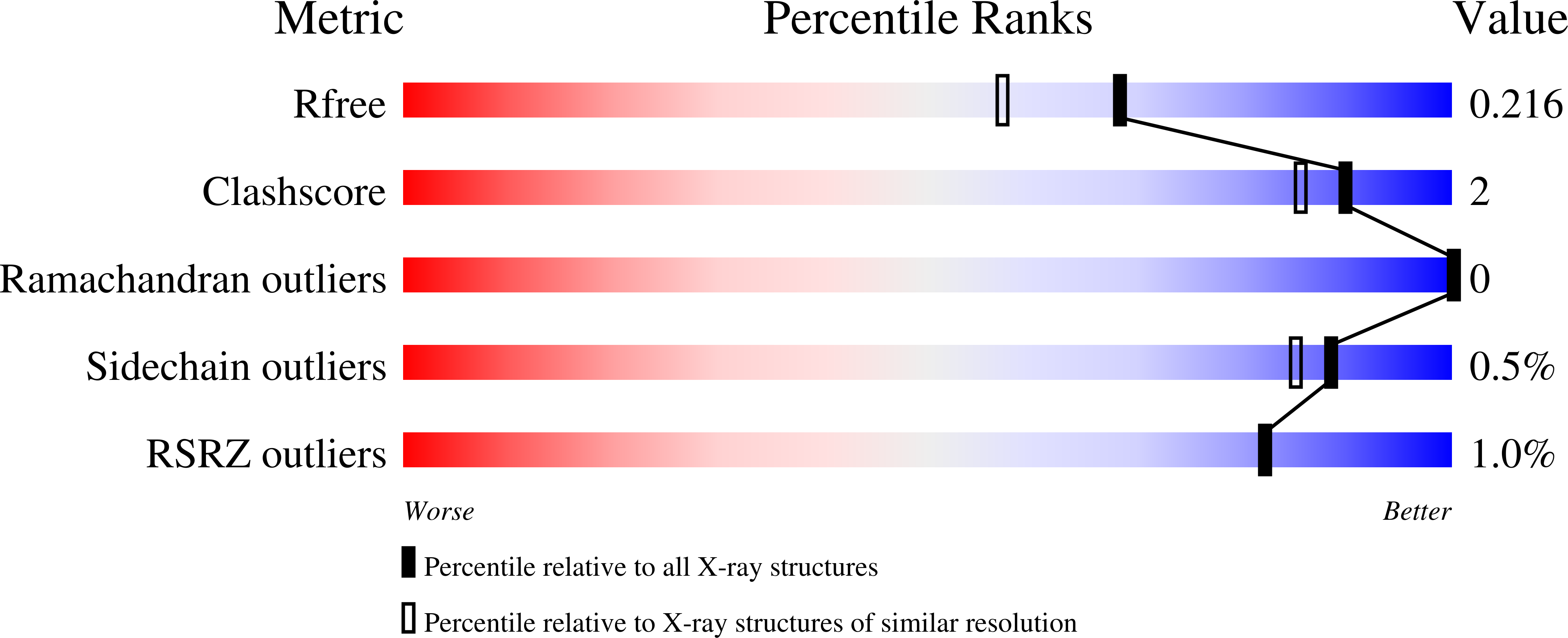
6UEH - PubMed Abstract:
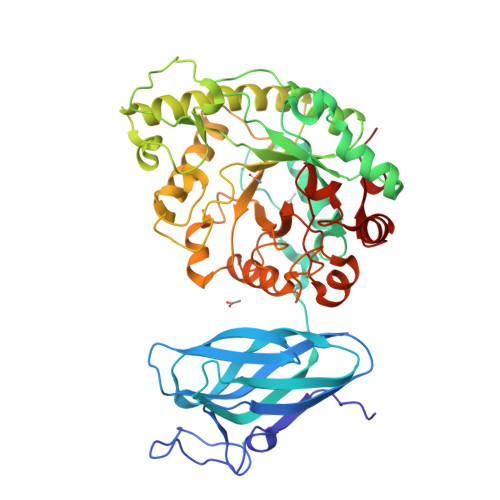
β-Mannanases from the glycoside hydrolase 26 (GH26) family are retaining hydrolases that are active on complex heteromannans and whose genes are abundant in rumen metagenomes and metatranscriptomes. These enzymes can exhibit distinct modes of substrate recognition and are often fused to carbohydrate-binding modules (CBMs), resulting in a molecular puzzle of mechanisms governing substrate preference and mode of action that has not yet been pieced together. In this study, we recovered a novel GH26 enzyme with a CBM35 module linked to its N terminus (CrMan26) from a cattle rumen metatranscriptome. CrMan26 exhibited a preference for galactomannan as substrate and the crystal structure of the full-length protein at 1.85 Å resolution revealed a unique orientation of the ancillary domain relative to the catalytic interface, strategically positioning a surface aromatic cluster of the ancillary domain as an extension of the substrate-binding cleft, contributing to galactomannan preference. Moreover, systematic investigation of nonconserved residues in the catalytic interface unveiled that residues Tyr 195 (-3 subsite) and Trp 234 (-5 subsite) from distal negative subsites have a key role in galactomannan preference. These results indicate a novel and complex mechanism for substrate recognition involving spatially remote motifs, distal negative subsites from the catalytic domain, and a surface-associated aromatic cluster from the ancillary domain. These findings expand our molecular understanding of the mechanisms of substrate binding and recognition in the GH26 family and shed light on how some CBMs and their respective orientation can contribute to substrate preference.
Organizational Affiliation:
Brazilian Biorenewables National Laboratory (LNBR), Brazilian Center for Research in Energy and Materials (CNPEM), Campinas, São Paulo 13083-970, Brazil.