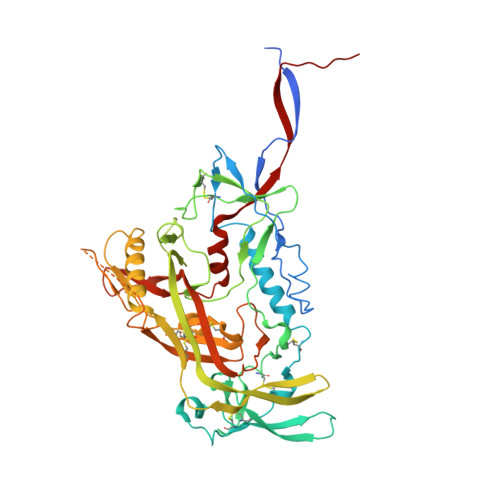
Neutralization-guided design of HIV-1 envelope trimers with high affinity for the unmutated common ancester of CH235 lineage CD4bs broadly neutralizing antibodies.
LaBranche, C.C., Henderson, R., Hsu, A., Behrens, S., Chen, X., Zhou, T., Wiehe, K., Saunders, K.O., Alam, S.M., Bonsignori, M., Borgnia, M.J., Sattentau, Q.J., Eaton, A., Greene, K., Gao, H., Liao, H.X., Williams, W.B., Peacock, J., Tang, H., Perez, L.G., Edwards, R.J., Kepler, T.B., Korber, B.T., Kwong, P.D., Mascola, J.R., Acharya, P., Haynes, B.F., Montefiori, D.C.(2019) PLoS Pathog 15: e1008026-e1008026
- PubMed: 31527908
- DOI: https://doi.org/10.1371/journal.ppat.1008026
- Primary Citation of Related Structures:
6UDA - PubMed Abstract:
The CD4 binding site (CD4bs) of the HIV-1 envelope glycoprotein is susceptible to multiple lineages of broadly neutralizing antibodies (bnAbs) that are attractive to elicit with vaccines. The CH235 lineage (VH1-46) of CD4bs bnAbs is particularly attractive because the most mature members neutralize 90% of circulating strains, do not possess long HCDR3 regions, and do not contain insertions and deletions that may be difficult to induce. We used virus neutralization to measure the interaction of CH235 unmutated common ancestor (CH235 UCA) with functional Env trimers on infectious virions to guide immunogen design for this bnAb lineage. Two Env mutations were identified, one in loop D (N279K) and another in V5 (G458Y), that acted synergistically to render autologous CH505 transmitted/founder virus susceptible to neutralization by CH235 UCA. Man5-enriched N-glycans provided additional synergy for neutralization. CH235 UCA bound with nanomolar affinity to corresponding soluble native-like Env trimers as candidate immunogens. A cryo-EM structure of CH235 UCA bound to Man5-enriched CH505.N279K.G458Y.SOSIP.664 revealed interactions of the antibody light chain complementarity determining region 3 (CDR L3) with the engineered Env loops D and V5. These results demonstrate that virus neutralization can directly inform vaccine design and suggest a germline targeting and reverse engineering strategy to initiate and mature the CH235 bnAb lineage.
- Department of Surgery, Duke University Medical Center, Durham, NC, United States of America.
Organizational Affiliation: