Revealing the mechanism of repressor inactivation during switching of a temperate bacteriophage.
Rasmussen, K.K., Palencia, A., Varming, A.K., El-Wali, H., Boeri Erba, E., Blackledge, M., Hammer, K., Herrmann, T., Kilstrup, M., Lo Leggio, L., Jensen, M.R.(2020) Proc Natl Acad Sci U S A 117: 20576-20585
- PubMed: 32788352
- DOI: https://doi.org/10.1073/pnas.2005218117
- Primary Citation of Related Structures:
6TO6, 6TRI - PubMed Abstract:
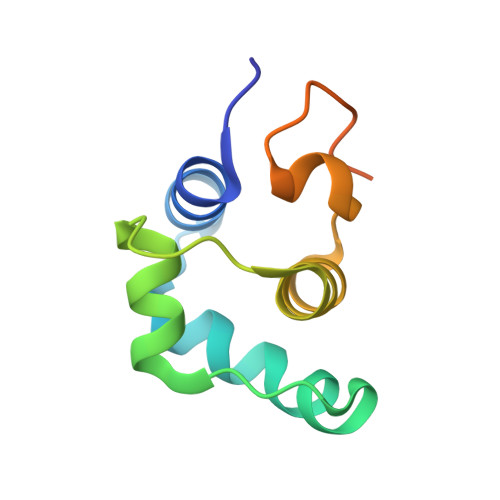
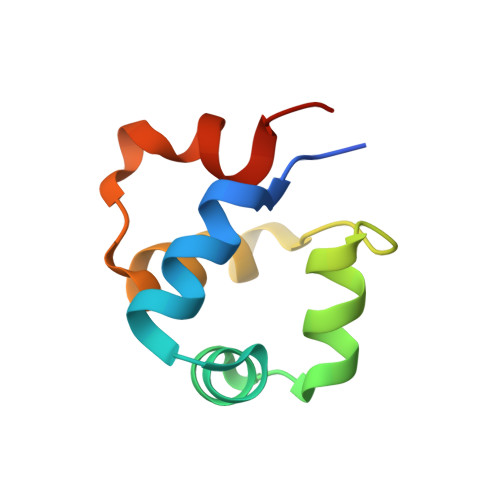
Temperate bacteriophages can enter one of two life cycles following infection of a sensitive host: the lysogenic or the lytic life cycle. The choice between the two alternative life cycles is dependent upon a tight regulation of promoters and their cognate regulatory proteins within the phage genome. We investigated the genetic switch of TP901-1, a bacteriophage of Lactococcus lactis , controlled by the CI repressor and the modulator of repression (MOR) antirepressor and their interactions with DNA. We determined the solution structure of MOR, and we solved the crystal structure of MOR in complex with the N-terminal domain of CI, revealing the structural basis of MOR inhibition of CI binding to the DNA operator sites. 15 N NMR Carr-Purcell-Meiboom-Gill (CPMG) relaxation dispersion and rotating frame R 1ρ measurements demonstrate that MOR displays molecular recognition dynamics on two different time scales involving a repacking of aromatic residues at the interface with CI. Mutations in the CI:MOR binding interface impair complex formation in vitro, and when introduced in vivo, the bacteriophage switch is unable to choose the lytic life cycle showing that the CI:MOR complex is essential for proper functioning of the genetic switch. On the basis of sequence alignments, we show that the structural features of the MOR:CI complex are likely conserved among a larger family of bacteriophages from human pathogens implicated in transfer of antibiotic resistance.
- Department of Chemistry, University of Copenhagen, DK-2100 Copenhagen, Denmark.
Organizational Affiliation: