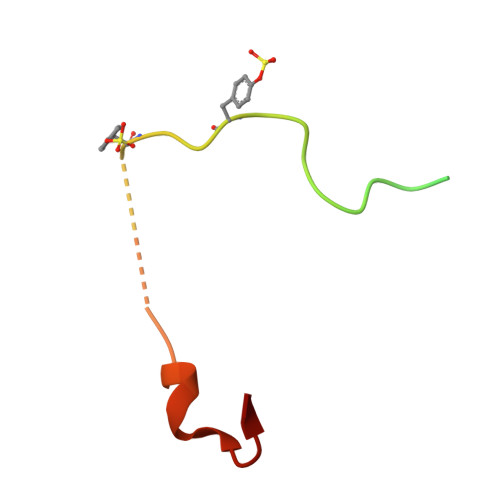
Sulfotyrosine-Mediated Recognition of Human Thrombin by a Tsetse Fly Anticoagulant Mimics Physiological Substrates.
Calisto, B.M., Ripoll-Rozada, J., Dowman, L.J., Franck, C., Agten, S.M., Parker, B.L., Veloso, R.C., Vale, N., Gomes, P., de Sanctis, D., Payne, R.J., Pereira, P.J.B.(2021) Cell Chem Biol 28: 26
- PubMed: 33096052
- DOI: https://doi.org/10.1016/j.chembiol.2020.10.002
- Primary Citation of Related Structures:
6TKG, 6TKH, 6TKI, 6TKJ, 6TKL - PubMed Abstract:
Despite possessing only 32 residues, the tsetse thrombin inhibitor (TTI) is among the most potent anticoagulants described, with sub-picomolar inhibitory activity against thrombin. Unexpectedly, TTI isolated from the fly is 2000-fold more active and 180 Da heavier than synthetic and recombinant variants. We predicted the presence of a tyrosine O-sulfate post-translational modification of TTI, prompting us to investigate the effect of the modification on anticoagulant activity. A combination of chemical synthesis and functional assays was used to reveal that sulfation significantly improved the inhibitory activity of TTI against thrombin. Using X-ray crystallography, we show that the N-terminal sulfated segment of TTI binds the basic exosite II of thrombin, establishing interactions similar to those of physiologic substrates, while the C-terminal segment abolishes the catalytic activity of thrombin. This non-canonical mode of inhibition, coupled with its potency and small size, makes TTI an attractive scaffold for the design of novel antithrombotics.
- ESRF - The European Synchrotron, Structural Biology Group, 38000 Grenoble, France; ALBA Synchrotron, 08290 Cerdanyola del Vallès, Spain.
Organizational Affiliation: