Structure of complete Pol II-DSIF-PAF-SPT6 transcription complex reveals RTF1 allosteric activation.
Vos, S.M., Farnung, L., Linden, A., Urlaub, H., Cramer, P.(2020) Nat Struct Mol Biol 27: 668-677
- PubMed: 32541898
- DOI: https://doi.org/10.1038/s41594-020-0437-1
- Primary Citation of Related Structures:
6TED - PubMed Abstract:
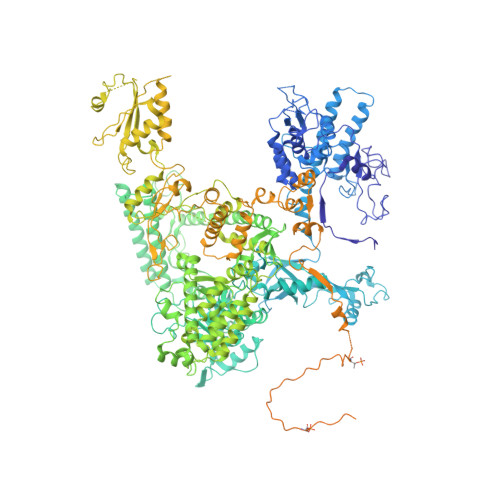
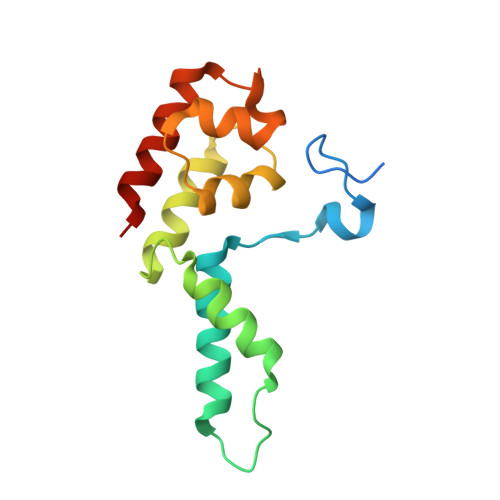
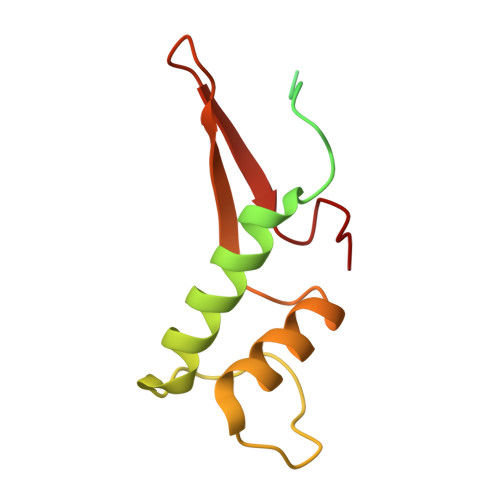
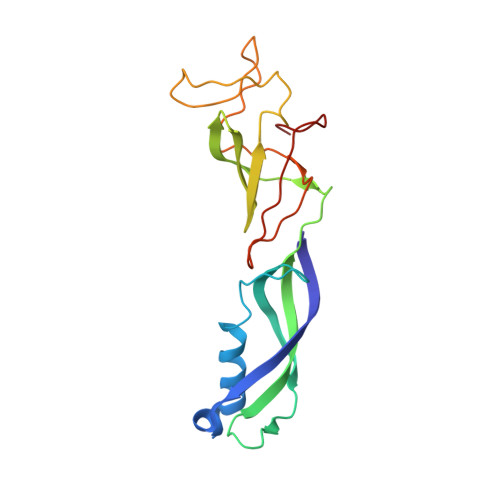
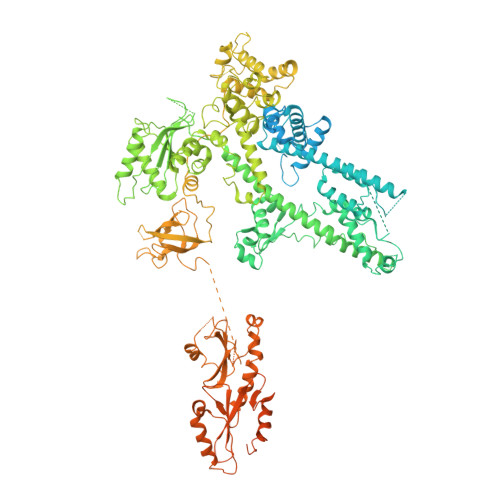
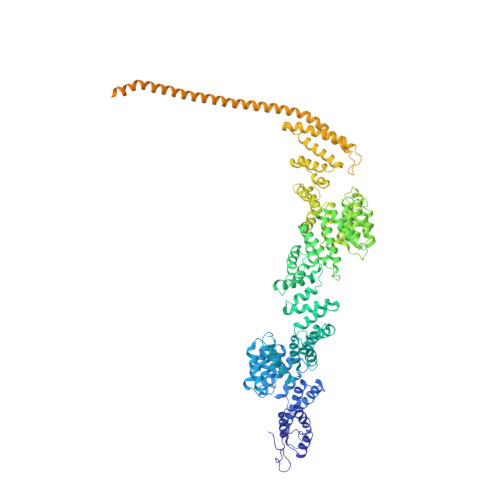
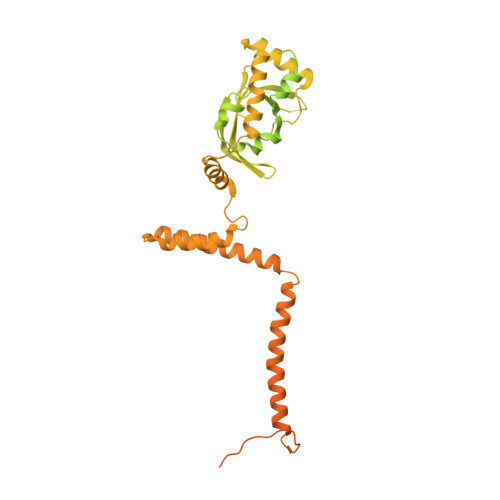
Transcription by RNA polymerase II (Pol II) is carried out by an elongation complex. We previously reported an activated porcine Pol II elongation complex, EC*, encompassing the human elongation factors DSIF, PAF1 complex (PAF) and SPT6. Here we report the cryo-EM structure of the complete EC* that contains RTF1, a dissociable PAF subunit critical for chromatin transcription. The RTF1 Plus3 domain associates with Pol II subunit RPB12 and the phosphorylated C-terminal region of DSIF subunit SPT5. RTF1 also forms four α-helices that extend from the Plus3 domain along the Pol II protrusion and RPB10 to the polymerase funnel. The C-terminal 'fastener' helix retains PAF and is followed by a 'latch' that reaches the end of the bridge helix, a flexible element of the Pol II active site. RTF1 strongly stimulates Pol II elongation, and this requires the latch, possibly suggesting that RTF1 activates transcription allosterically by influencing Pol II translocation.
- Max Planck Institute for Biophysical Chemistry, Department of Molecular Biology, Göttingen, Germany.
Organizational Affiliation: