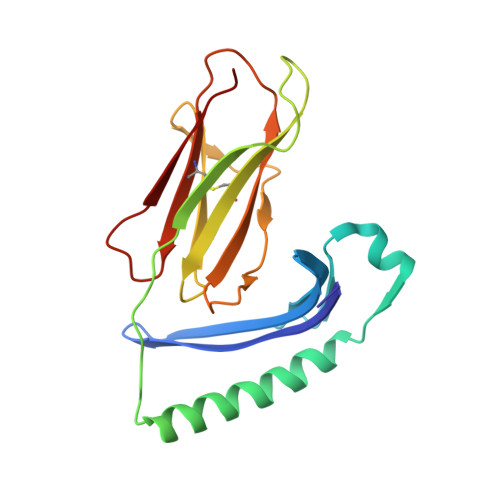
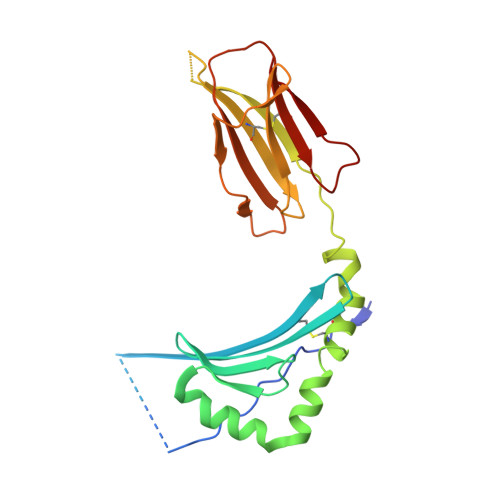
The dominantly expressed class II molecule from a resistant MHC haplotype presents only a few Marek's disease virus peptides by using an unprecedented binding motif.
Halabi, S., Ghosh, M., Stevanovic, S., Rammensee, H.G., Bertzbach, L.D., Kaufer, B.B., Moncrieffe, M.C., Kaspers, B., Hartle, S., Kaufman, J.(2021) PLoS Biol 19: e3001057-e3001057
- PubMed: 33901176
- DOI: https://doi.org/10.1371/journal.pbio.3001057
- Primary Citation of Related Structures:
6T3Y - PubMed Abstract:
Viral diseases pose major threats to humans and other animals, including the billions of chickens that are an important food source as well as a public health concern due to zoonotic pathogens. Unlike humans and other typical mammals, the major histocompatibility complex (MHC) of chickens can confer decisive resistance or susceptibility to many viral diseases. An iconic example is Marek's disease, caused by an oncogenic herpesvirus with over 100 genes. Classical MHC class I and class II molecules present antigenic peptides to T lymphocytes, and it has been hard to understand how such MHC molecules could be involved in susceptibility to Marek's disease, given the potential number of peptides from over 100 genes. We used a new in vitro infection system and immunopeptidomics to determine peptide motifs for the 2 class II molecules expressed by the MHC haplotype B2, which is known to confer resistance to Marek's disease. Surprisingly, we found that the vast majority of viral peptide epitopes presented by chicken class II molecules arise from only 4 viral genes, nearly all having the peptide motif for BL2*02, the dominantly expressed class II molecule in chickens. We expressed BL2*02 linked to several Marek's disease virus (MDV) peptides and determined one X-ray crystal structure, showing how a single small amino acid in the binding site causes a crinkle in the peptide, leading to a core binding peptide of 10 amino acids, compared to the 9 amino acids in all other reported class II molecules. The limited number of potential T cell epitopes from such a complex virus can explain the differential MHC-determined resistance to MDV, but raises questions of mechanism and opportunities for vaccine targets in this important food species, as well as providing a basis for understanding class II molecules in other species including humans.
- University of Cambridge, Department of Pathology, Cambridge, United Kingdom.
Organizational Affiliation: